Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Mazzei, L., Cianci, M., Contaldo, U., Musiani, F., Ciurli, S.(2017) Biochemistry 56: 5391-5404
- PubMed: 28857549
- DOI: https://doi.org/10.1021/acs.biochem.7b00750
- Primary Citation of Related Structures:
5OL4 - PubMed Abstract:
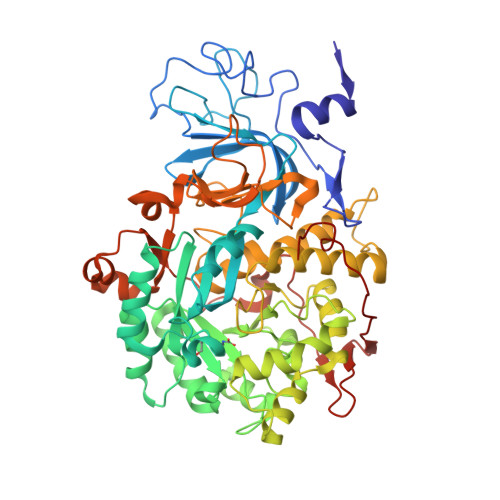
The nickel-dependent enzyme urease is a virulence factor for a large number of pathogenic and antibiotic-resistant bacteria, as well as a negative factor for the efficiency of soil nitrogen fertilization for crop production. The use of urease inhibitors to offset these effects requires knowledge, at a molecular level, of their mode of action. The 1.28 Å resolution structure of the enzyme-inhibitor complex obtained upon incubation of Sporosarcina pasteurii urease with N-(n-butyl)thiophosphoric triamide (NBPT), a molecule largely utilized in agriculture, reveals the presence of the monoamidothiophosphoric acid (MATP) moiety, obtained upon enzymatic hydrolysis of the diamide derivative of NBPT (NBPD) to yield n-butyl amine. MATP is bound to the two Ni(II) ions in the active site of urease using a μ 2 -bridging O atom and terminally bound O and NH 2 groups, with the S atom of the thiophosphoric amide pointing away from the metal center. The mobile flap modulating the size of the active site cavity is found in the closed conformation. Docking calculations suggest that the interaction between urease in the open flap conformation and NBPD involves a role for the conserved αArg339 in capturing and orienting the inhibitor prior to flap closure. Calorimetric and spectrophotometric determinations of the kinetic parameters of this inhibition indicate the occurrence of a reversible slow inhibition mode of action, characterized, for both bacterial and plant ureases, by a very small value of the dissociation constant of the urease-MATP complex. No need to convert NBPT to its oxo derivative NBPTO, as previously proposed, is necessary for urease inhibition.
- Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna , Bologna, Italy.
Organizational Affiliation: