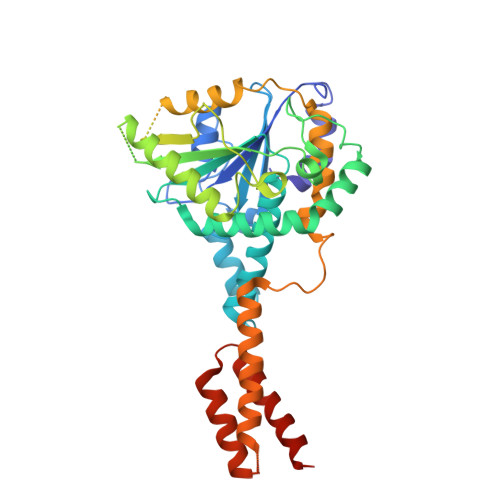
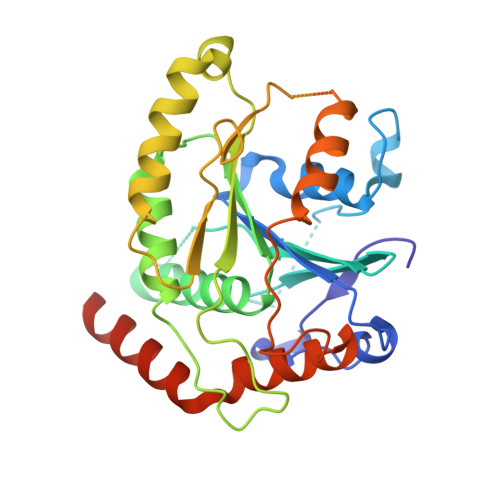
Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
Li, L., Lingaraju, M., Basquin, C., Basquin, J., Conti, E.(2017) RNA 23: 1028-1034
- PubMed: 28389433
- DOI: https://doi.org/10.1261/rna.061200.117
- Primary Citation of Related Structures:
5NKK, 5NKM - PubMed Abstract:
Nonsense-mediated mRNA decay (NMD) is a eukaryotic mRNA degradation pathway involved in surveillance and post-transcriptional regulation, and executed by the concerted action of several trans -acting factors. The SMG1 kinase is an essential NMD factor in metazoans and is associated with two recently identified and yet poorly characterized proteins, SMG8 and SMG9. We determined the 2.5 Å resolution crystal structure of a SMG8-SMG9 core complex from C. elegans We found that SMG8-SMG9 is a G-domain heterodimer with architectural similarities to the dynamin-like family of GTPases such as Atlastin and GBP1. The SMG8-SMG9 heterodimer forms in the absence of nucleotides, with interactions conserved from worms to humans. Nucleotide binding occurs at the G domain of SMG9 but not of SMG8. Fitting the GDP-bound SMG8-SMG9 structure in EM densities of the human SMG1-SMG8-SMG9 complex raises the possibility that the nucleotide site of SMG9 faces SMG1 and could impact the kinase conformation and/or regulation.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, D-82152 Martinsried, Germany.
Organizational Affiliation: