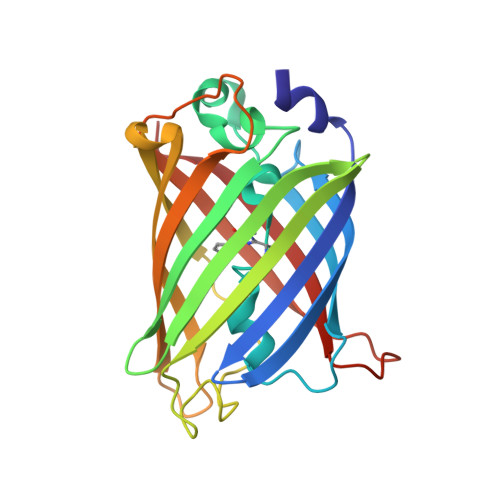
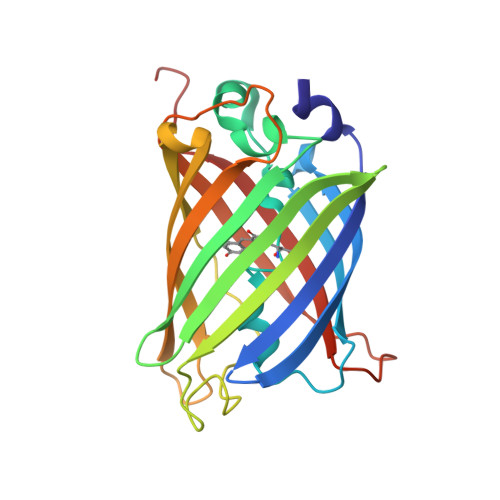
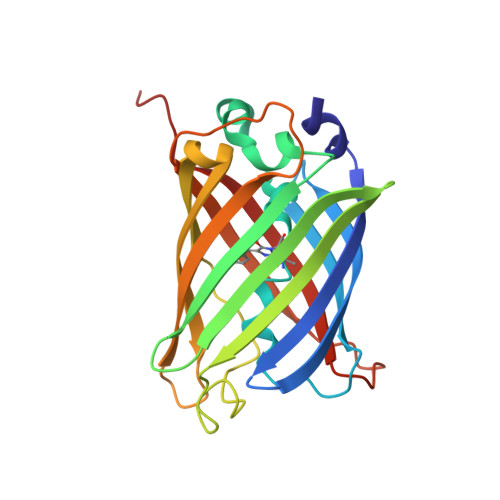
Association of Fluorescent Protein Pairs and Its Significant Impact on Fluorescence and Energy Transfer
Pope, J.R., Johnson, R.L., Jamieson, W.D., Worthy, H.L., Kailasam, S., Ahmed, R.D., Taban, I., Auhim, H.S., Watkins, D.W., Rizkallah, P.J., Castell, O.K., Jones, D.D.(2020) Adv Sci (Weinh)