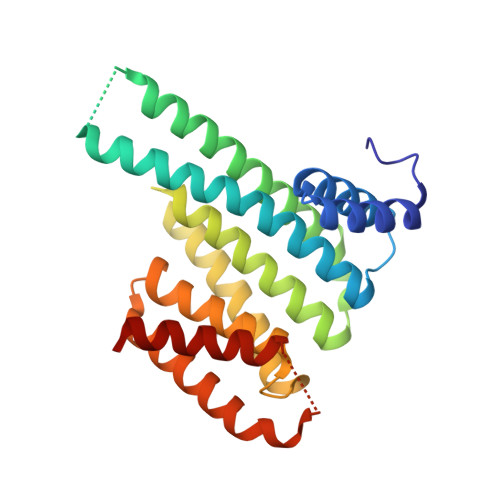
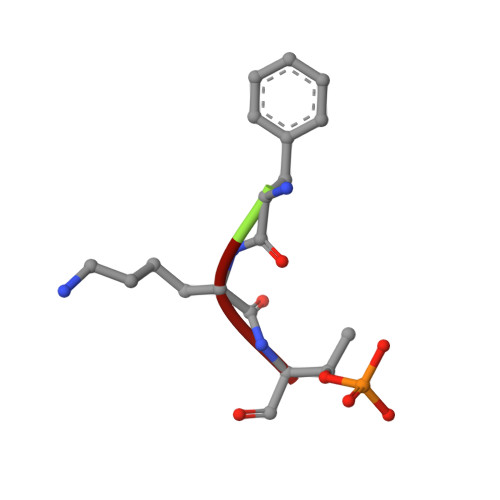
Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
Doveston, R.G., Kuusk, A., Andrei, S.A., Leysen, S., Cao, Q., Castaldi, M.P., Hendricks, A., Brunsveld, L., Chen, H., Boyd, H., Ottmann, C.(2017) FEBS Lett 591: 2449-2457
- PubMed: 28640363
- DOI: https://doi.org/10.1002/1873-3468.12723
- Primary Citation of Related Structures:
5MHC, 5MOC, 5MXO - PubMed Abstract:
14-3-3 proteins are positive regulators of the tumor suppressor p53, the mutation of which is implicated in many human cancers. Current strategies for targeting of p53 involve restoration of wild-type function or inhibition of the interaction with MDM2, its key negative regulator. Despite the efficacy of these strategies, the alternate approach of stabilizing the interaction of p53 with positive regulators and, thus, enhancing tumor suppressor activity, has not been explored. Here, we report the first example of small-molecule stabilization of the 14-3-3 - p53 protein-protein interaction (PPI) and demonstrate the potential of this approach as a therapeutic modality. We also observed a disconnect between biophysical and crystallographic data in the presence of a stabilizing molecule, which is unusual in 14-3-3 PPIs.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, The Netherlands.
Organizational Affiliation: