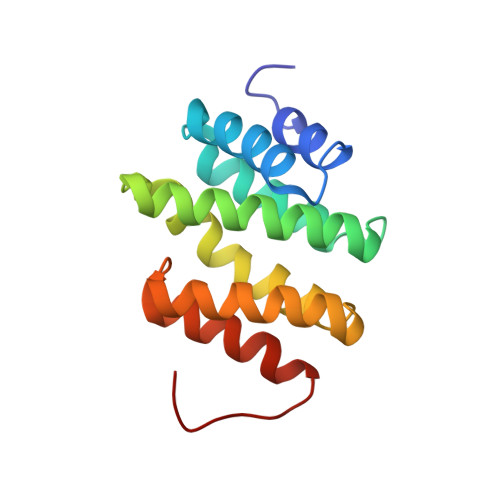
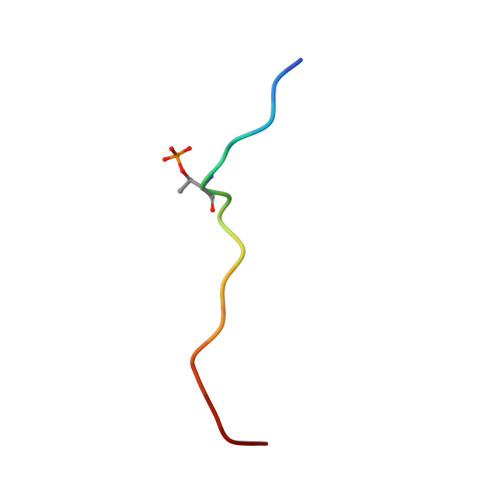
Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p.
Jasnovidova, O., Krejcikova, M., Kubicek, K., Stefl, R.(2017) EMBO Rep 18: 906-913
- PubMed: 28468956
- DOI: https://doi.org/10.15252/embr.201643723
- Primary Citation of Related Structures:
5LVF - PubMed Abstract:
Phosphorylation patterns of the C-terminal domain (CTD) of largest subunit of RNA polymerase II (called the CTD code) orchestrate the recruitment of RNA processing and transcription factors. Recent studies showed that not only serines and tyrosines but also threonines of the CTD can be phosphorylated with a number of functional consequences, including the interaction with yeast transcription termination factor, Rtt103p. Here, we report the solution structure of the Rtt103p CTD-interacting domain (CID) bound to Thr4 phosphorylated CTD, a poorly understood letter of the CTD code. The structure reveals a direct recognition of the phospho-Thr4 mark by Rtt103p CID and extensive interactions involving residues from three repeats of the CTD heptad. Intriguingly, Rtt103p's CID binds equally well Thr4 and Ser2 phosphorylated CTD A doubly phosphorylated CTD at Ser2 and Thr4 diminishes its binding affinity due to electrostatic repulsion. Our structural data suggest that the recruitment of a CID-containing CTD-binding factor may be coded by more than one letter of the CTD code.
- CEITEC - Central European Institute of Technology, Masaryk University, Brno, Czech Republic olga.jasnovidova@ceitec.muni.cz richard.stefl@ceitec.muni.cz.
Organizational Affiliation: