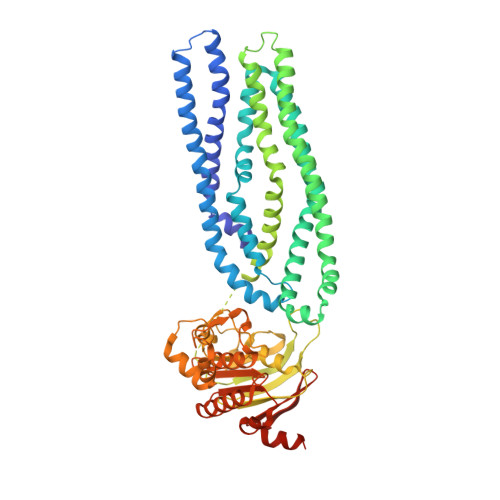
Structure of a Type-1 Secretion System ABC Transporter.
Morgan, J.L., Acheson, J.F., Zimmer, J.(2017) Structure 25: 522-529
- PubMed: 28216041
- DOI: https://doi.org/10.1016/j.str.2017.01.010
- Primary Citation of Related Structures:
5L22 - PubMed Abstract:
Type-1 secretion systems (T1SSs) represent a widespread mode of protein secretion across the cell envelope in Gram-negative bacteria. The T1SS is composed of an inner-membrane ABC transporter, a periplasmic membrane-fusion protein, and an outer-membrane porin. These three components assemble into a complex spanning both membranes and providing a conduit for the translocation of unfolded polypeptides. We show that ATP hydrolysis and assembly of the entire T1SS complex is necessary for protein secretion. Furthermore, we present a 3.15-Å crystal structure of AaPrtD, the ABC transporter found in the Aquifex aeolicus T1SS. The structure suggests a substrate entry window just above the transporter's nucleotide binding domains. In addition, highly kinked transmembrane helices, which frame a narrow channel not observed in canonical peptide transporters, are likely involved in substrate translocation. Overall, the AaPrtD structure supports a polypeptide transport mechanism distinct from alternating access.
- Department of Molecular Physiology and Biological Physics, University of Virginia School of Medicine, 480 Ray C. Hunt Drive, Charlottesville, VA 22908, USA. Electronic address: jlm2qp@virginia.edu.
Organizational Affiliation: