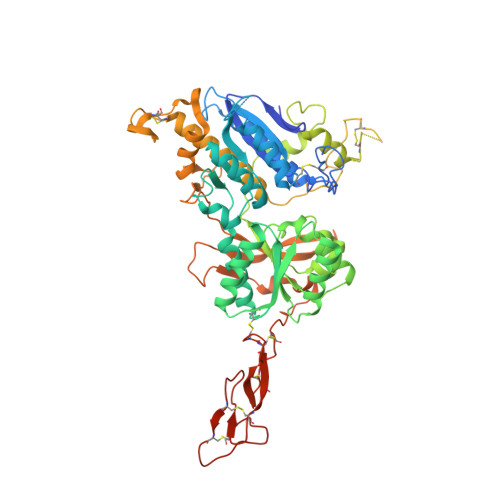
Structural mechanism of ligand activation in human calcium-sensing receptor.
Geng, Y., Mosyak, L., Kurinov, I., Zuo, H., Sturchler, E., Cheng, T.C., Subramanyam, P., Brown, A.P., Brennan, S.C., Mun, H.C., Bush, M., Chen, Y., Nguyen, T.X., Cao, B., Chang, D.D., Quick, M., Conigrave, A.D., Colecraft, H.M., McDonald, P., Fan, Q.R.(2016) Elife 5
- PubMed: 27434672
- DOI: https://doi.org/10.7554/eLife.13662
- Primary Citation of Related Structures:
5K5S, 5K5T - PubMed Abstract:
Human calcium-sensing receptor (CaSR) is a G-protein-coupled receptor (GPCR) that maintains extracellular Ca(2+) homeostasis through the regulation of parathyroid hormone secretion. It functions as a disulfide-tethered homodimer composed of three main domains, the Venus Flytrap module, cysteine-rich domain, and seven-helix transmembrane region. Here, we present the crystal structures of the entire extracellular domain of CaSR in the resting and active conformations. We provide direct evidence that L-amino acids are agonists of the receptor. In the active structure, L-Trp occupies the orthosteric agonist-binding site at the interdomain cleft and is primarily responsible for inducing extracellular domain closure to initiate receptor activation. Our structures reveal multiple binding sites for Ca(2+) and PO4(3-) ions. Both ions are crucial for structural integrity of the receptor. While Ca(2+) ions stabilize the active state, PO4(3-) ions reinforce the inactive conformation. The activation mechanism of CaSR involves the formation of a novel dimer interface between subunits.
- Department of Pharmacology, Columbia University, New York, United States.
Organizational Affiliation: