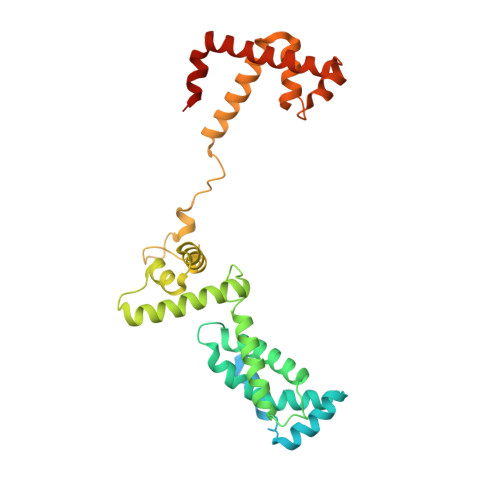
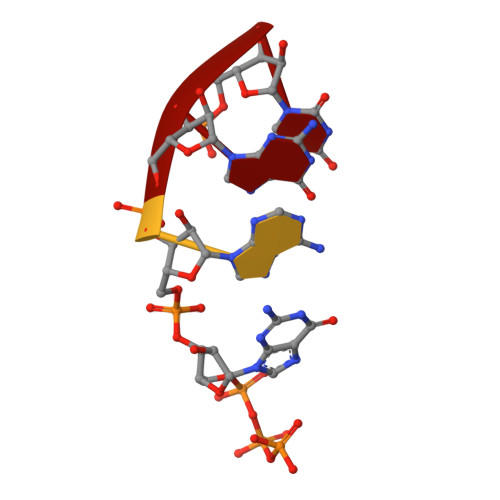
Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Liu, B., Zuo, Y., Steitz, T.A.(2016) Proc Natl Acad Sci U S A 113: 4051-4056
- PubMed: 27035955
- DOI: https://doi.org/10.1073/pnas.1520555113
- Primary Citation of Related Structures:
5IPL, 5IPM, 5IPN - PubMed Abstract:
In bacteria, multiple σ factors compete to associate with the RNA polymerase (RNAP) core enzyme to form a holoenzyme that is required for promoter recognition. During transcription initiation RNAP remains associated with the upstream promoter DNA via sequence-specific interactions between the σ factor and the promoter DNA while moving downstream for RNA synthesis. As RNA polymerase repetitively adds nucleotides to the 3'-end of the RNA, a pyrophosphate ion is generated after each nucleotide incorporation. It is currently unknown how the release of pyrophosphate affects transcription. Here we report the crystal structures of E coli transcription initiation complexes (TICs) containing the stress-responsive σ(S) factor, a de novo synthesized RNA oligonucleotide, and a complete transcription bubble (σ(S)-TIC) at about 3.9-Å resolution. The structures show the 3D topology of the σ(S) factor and how it recognizes the promoter DNA, including likely specific interactions with the template-strand residues of the -10 element. In addition, σ(S)-TIC structures display a highly stressed pretranslocated initiation complex that traps a pyrophosphate at the active site that remains closed. The position of the pyrophosphate and the unusual phosphodiester linkage between the two terminal RNA residues suggest an unfinished nucleotide-addition reaction that is likely at equilibrium between nucleotide addition and pyrophosphorolysis. Although these σ(S)-TIC crystals are enzymatically active, they are slow in nucleotide addition, as suggested by an NTP soaking experiment. Pyrophosphate release completes the nucleotide addition reaction and is associated with extensive conformational changes around the secondary channel but causes neither active site opening nor transcript translocation.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520;
Organizational Affiliation: