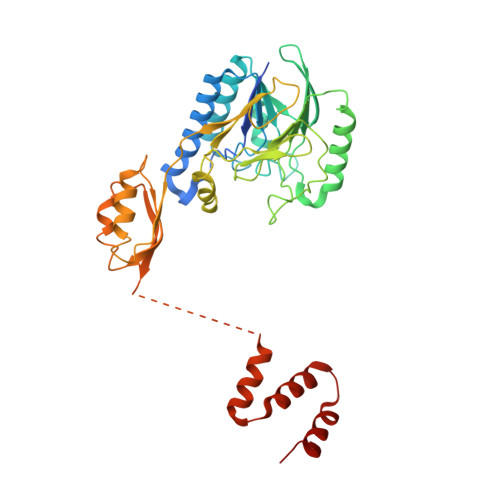
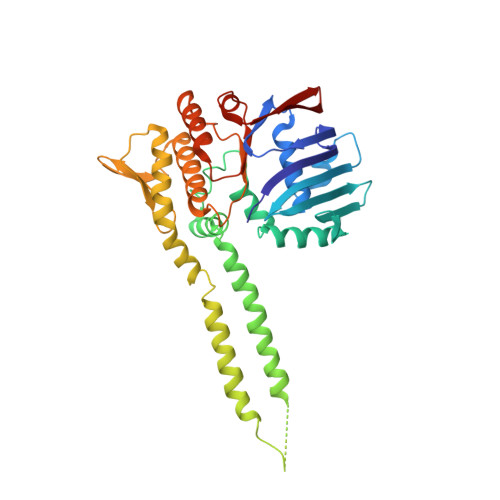
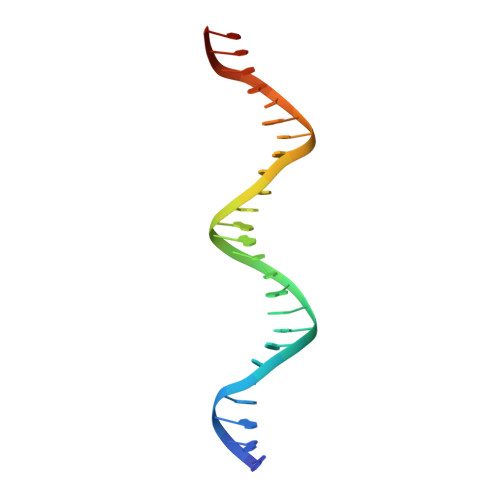
ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Liu, Y., Sung, S., Kim, Y., Li, F., Gwon, G., Jo, A., Kim, A.K., Kim, T., Song, O.K., Lee, S.E., Cho, Y.(2016) EMBO J 35: 743-758
- PubMed: 26717941
- DOI: https://doi.org/10.15252/embj.201592462
- Primary Citation of Related Structures:
5DNY, 5F3W - PubMed Abstract:
ATP-dependent DNA end recognition and nucleolytic processing are central functions of the Mre11/Rad50 (MR) complex in DNA double-strand break repair. However, it is still unclear how ATP binding and hydrolysis primes the MR function and regulates repair pathway choice in cells. Here,Methanococcus jannaschii MR-ATPγS-DNA structure reveals that the partly deformed DNA runs symmetrically across central groove between two ATPγS-bound Rad50 nucleotide-binding domains. Duplex DNA cannot access the Mre11 active site in the ATP-free full-length MR complex. ATP hydrolysis drives rotation of the nucleotide-binding domain and induces the DNA melting so that the substrate DNA can access Mre11. Our findings suggest that the ATP hydrolysis-driven conformational changes in both DNA and the MR complex coordinate the melting and endonuclease activity.
- Department of Life Sciences, Pohang University of Science and Technology, Pohang, South Korea.
Organizational Affiliation: