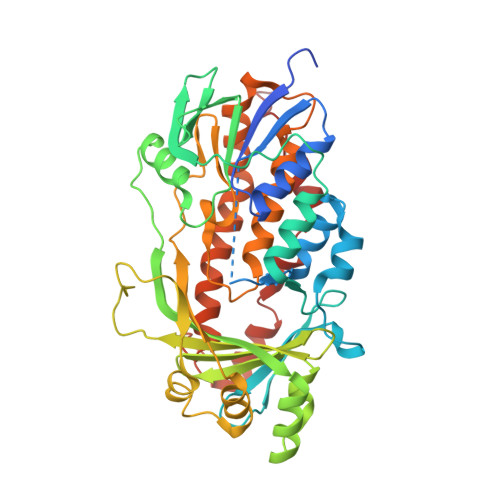
The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Uemura, T., Kita, A., Watanabe, Y., Adachi, M., Kuroki, R., Morimoto, Y.(2016) Biochem Biophys Res Commun 469: 158-163
- PubMed: 26616054
- DOI: https://doi.org/10.1016/j.bbrc.2015.11.087
- Primary Citation of Related Structures:
5EVY - PubMed Abstract:
The X-ray crystal structure of a salicylate hydroxylase from Pseudomonas putida S-1 complexed with coenzyme FAD has been determined to a resolution of 2.5 Å. Structural conservation with p- or m-hydroxybenzoate hydroxylase is very good throughout the topology, despite a low amino sequence identity of 20-40% between these three hydroxylases. Salicylate hydroxylase is composed of three distinct domains and includes FAD between domains I and II, which is accessible to solvent. In this study, which analyzes the tertiary structure of the enzyme, the unique reaction of salicylate, i.e. decarboxylative hydroxylation, and the structural roles of amino acids surrounding the substrate, are considered.
- Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan; Research Reactor Institute, Kyoto University, Kumatori, Osaka 590-0494, Japan.
Organizational Affiliation: