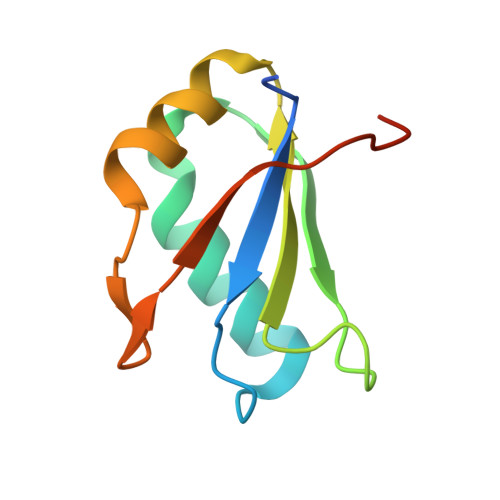
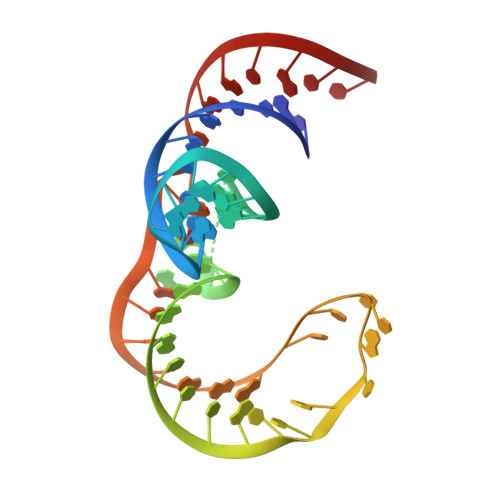
Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Ren, A., Xue, Y., Peselis, A., Serganov, A., Al-Hashimi, H.M., Patel, D.J.(2015) Cell Rep 13: 1800-1813
- PubMed: 26655897
- DOI: https://doi.org/10.1016/j.celrep.2015.10.062
- Primary Citation of Related Structures:
5DDO, 5DDP, 5DDQ, 5DDR - PubMed Abstract:
Naturally occurring L-glutamine riboswitches occur in cyanobacteria and marine metagenomes, where they reside upstream of genes involved in nitrogen metabolism. By combining X-ray, NMR, and MD, we characterized an L-glutamine-dependent conformational transition in the Synechococcus elongatus glutamine riboswitch from tuning fork to L-shaped alignment of stem segments. This transition generates an open ligand-binding pocket with L-glutamine selectivity enforced by Mg(2+)-mediated intermolecular interactions. The transition also stabilizes the P1 helix through a long-range "linchpin" Watson-Crick G-C pair-capping interaction, while melting a short helix below P1 potentially capable of modulating downstream readout. NMR data establish that the ligand-free glutamine riboswitch in Mg(2+) solution exists in a slow equilibrium between flexible tuning fork and a minor conformation, similar, but not identical, to the L-shaped bound conformation. We propose that an open ligand-binding pocket combined with a high conformational penalty for forming the ligand-bound state provide mechanisms for reducing binding affinity while retaining high selectivity.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: