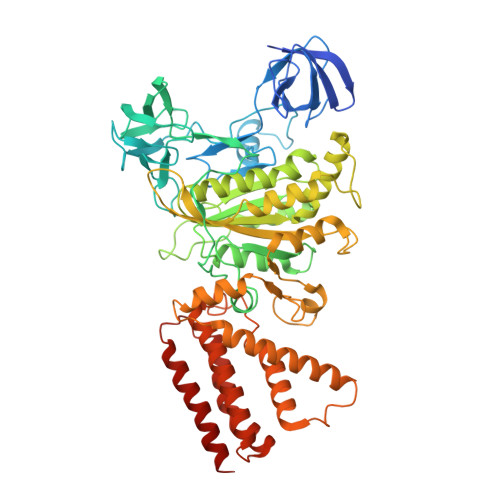
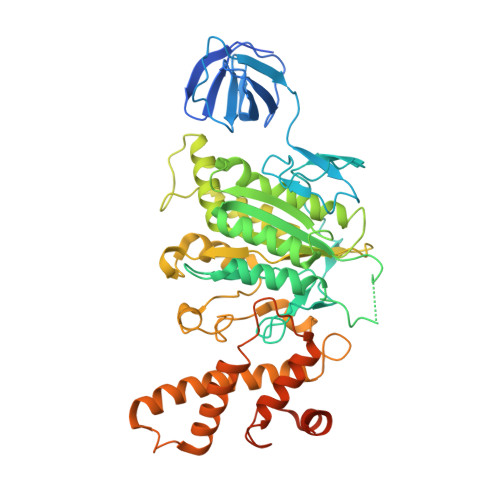
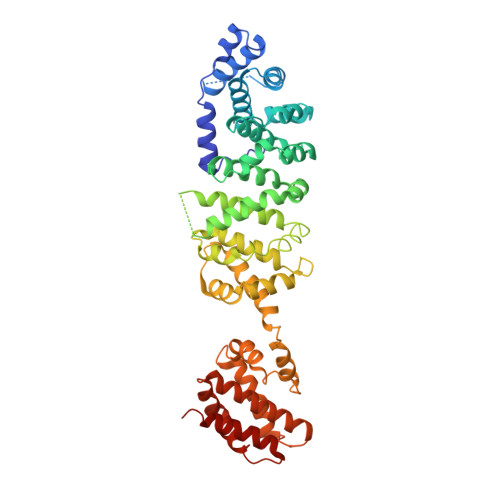
Crystal structure of yeast V1-ATPase in the autoinhibited state.
Oot, R.A., Kane, P.M., Berry, E.A., Wilkens, S.(2016) EMBO J 35: 1694-1706
- PubMed: 27295975
- DOI: https://doi.org/10.15252/embj.201593447
- Primary Citation of Related Structures:
5BW9, 5D80 - PubMed Abstract:
Vacuolar ATPases (V-ATPases) are essential proton pumps that acidify the lumen of subcellular organelles in all eukaryotic cells and the extracellular space in some tissues. V-ATPase activity is regulated by a unique mechanism referred to as reversible disassembly, wherein the soluble catalytic sector, V1, is released from the membrane and its MgATPase activity silenced. The crystal structure of yeast V1 presented here shows that activity silencing involves a large conformational change of subunit H, with its C-terminal domain rotating ~150° from a position near the membrane in holo V-ATPase to a position at the bottom of V1 near an open catalytic site. Together with biochemical data, the structure supports a mechanistic model wherein subunit H inhibits ATPase activity by stabilizing an open catalytic site that results in tight binding of inhibitory ADP at another site.
- Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, USA.
Organizational Affiliation: