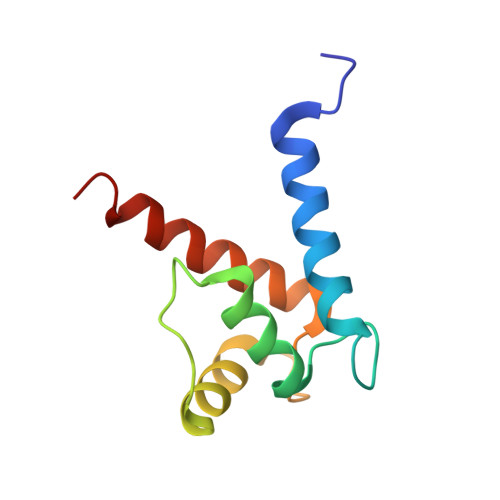
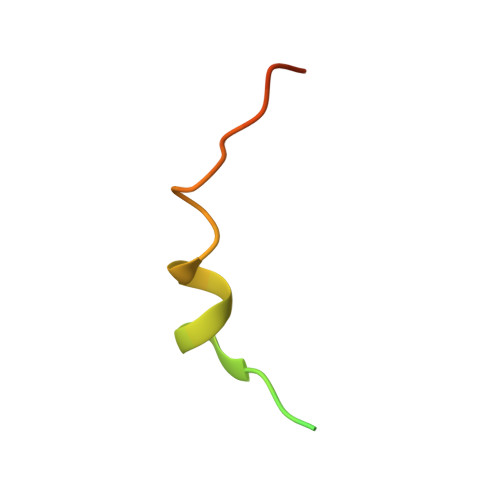
Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
Gogl, G., Alexa, A., Kiss, B., Katona, G., Kovacs, M., Bodor, A., Remenyi, A., Nyitray, L.(2016) J Biological Chem 291: 11-27
- PubMed: 26527685
- DOI: https://doi.org/10.1074/jbc.M115.684928
- Primary Citation of Related Structures:
5CSF, 5CSI, 5CSJ, 5CSN - PubMed Abstract:
Mitogen-activated protein kinases (MAPK) promote MAPK-activated protein kinase activation. In the MAPK pathway responsible for cell growth, ERK2 initiates the first phosphorylation event on RSK1, which is inhibited by Ca(2+)-binding S100 proteins in malignant melanomas. Here, we present a detailed in vitro biochemical and structural characterization of the S100B-RSK1 interaction. The Ca(2+)-dependent binding of S100B to the calcium/calmodulin-dependent protein kinase (CaMK)-type domain of RSK1 is reminiscent of the better known binding of calmodulin to CaMKII. Although S100B-RSK1 and the calmodulin-CAMKII system are clearly distinct functionally, they demonstrate how unrelated intracellular Ca(2+)-binding proteins could influence the activity of the CaMK domain-containing protein kinases. Our crystallographic, small angle x-ray scattering, and NMR analysis revealed that S100B forms a "fuzzy" complex with RSK1 peptide ligands. Based on fast-kinetics experiments, we conclude that the binding involves both conformation selection and induced fit steps. Knowledge of the structural basis of this interaction could facilitate therapeutic targeting of melanomas.
- From the Department of Biochemistry.
Organizational Affiliation: