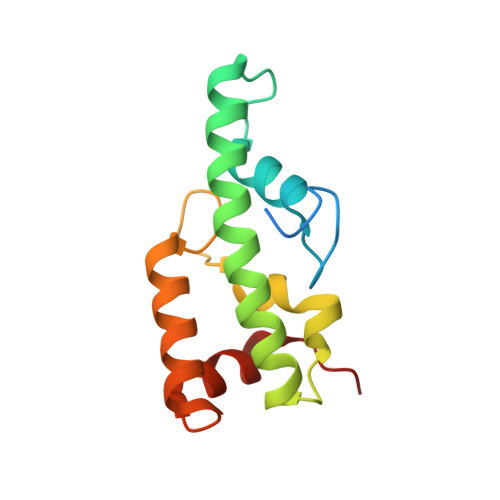
Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice
Wang, X., Guan, Z., Gong, Z., Yan, J., Yang, G., Liu, Y.G., Yin, P.(2018) Biochem Biophys Res Commun 501: 898-904
- PubMed: 29775612
- DOI: https://doi.org/10.1016/j.bbrc.2018.05.079
- Primary Citation of Related Structures:
5ZT3 - PubMed Abstract:
Plant cytoplasmic male sterility (CMS) is an important phenomenon and is widely utilized in hybrid crop breeding. The Wild Abortive CMS (CMS-WA), a well-known CMS type, has been successfully applied in the commercial production of hybrid rice seeds for more than 40 years. The CMS-WA causal gene WA352 encodes a novel transmenbrane protein and the interacts with the mitochondrial copper chaperone COX11, triggering reactive oxygen species production and resulting in male sterility in CMS-WA lines. However, the structure of WA352 is currently unknown, and the structural mechanism whereby WA352 perturbs COX11 function to cause CMS remains largely unknown. Here, we report the crystal structure of the C-terminal functional domain of WA352 at 1.3 Å resolution. This functional domain, consisting of five α helices, is spindle-shaped with a length of 42 Å, and a diameter of 28 Å. Notably, the absence of any structural similarity to a known protein structure suggests that the WA352 functional domain is a novel fold. In addition, surface conservation analysis and structural modeling of the WA352-COX11 complex revealed details about the WA352-COX11 interaction. Further structural analysis suggested that the WA352-COX11 interaction blocks the copper ion transportation activity of COX11, which is essential for the assembly of cytochrome c oxidase, resulting in male sterility in CMS-WA lines. Our study paves the way toward structural determination of the WA352-COX11 complex and provides new insight into the mechanism of plant CMS.
- National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Organizational Affiliation: