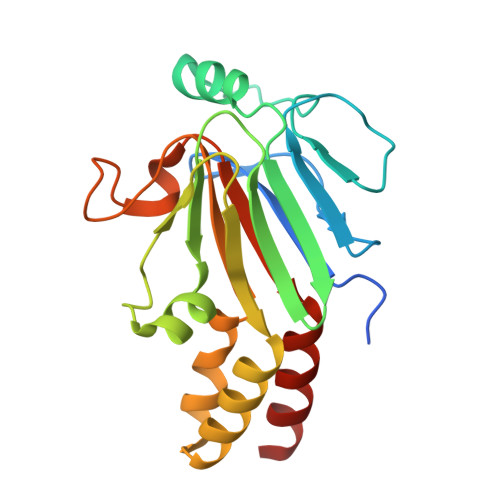
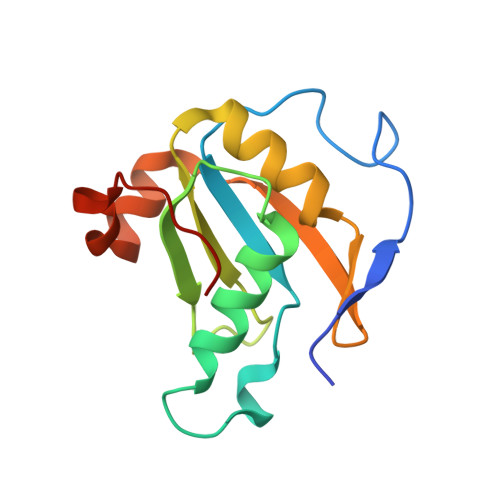
Structural basis for receptor-regulated SMAD recognition by MAN1
Miyazono, K.I., Ohno, Y., Wada, H., Ito, T., Fukatsu, Y., Kurisaki, A., Asashima, M., Tanokura, M.(2018) Nucleic Acids Res 46: 12139-12153
- PubMed: 30321401
- DOI: https://doi.org/10.1093/nar/gky925
- Primary Citation of Related Structures:
5ZOJ, 5ZOK - PubMed Abstract:
Receptor-regulated SMAD (R-SMAD: SMAD1, SMAD2, SMAD3, SMAD5 and SMAD8) proteins are key transcription factors of the transforming growth factor-β (TGF-β) superfamily of cytokines. MAN1, an integral protein of the inner nuclear membrane, is a SMAD cofactor that terminates TGF-β superfamily signals. Heterozygous loss-of-function mutations in MAN1 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. MAN1 interacts with MAD homology 2 (MH2) domains of R-SMAD proteins using its C-terminal U2AF homology motif (UHM) domain and UHM ligand motif (ULM) and facilitates R-SMAD dephosphorylation. Here, we report the structural basis for R-SMAD recognition by MAN1. The SMAD2-MAN1 and SMAD1-MAN1 complex structures show that an intramolecular UHM-ULM interaction of MAN1 forms a hydrophobic surface that interacts with a hydrophobic surface among the H2 helix, the strands β8 and β9, and the L3 loop of the MH2 domains of R-SMAD proteins. The complex structures also show the mechanism by which SMAD cofactors distinguish R-SMAD proteins that possess a highly conserved molecular surface.
- Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Organizational Affiliation: