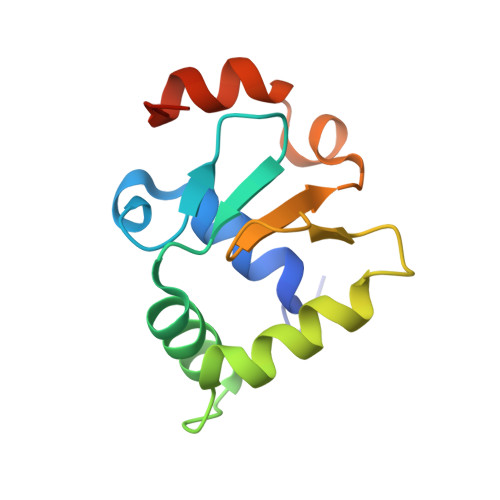
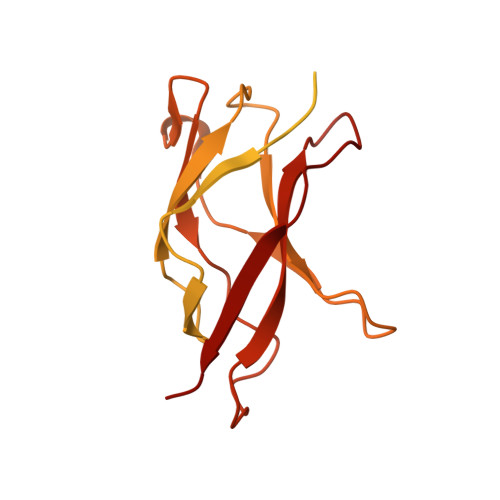
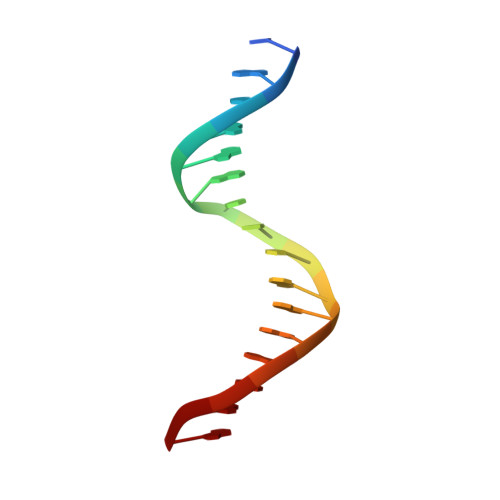
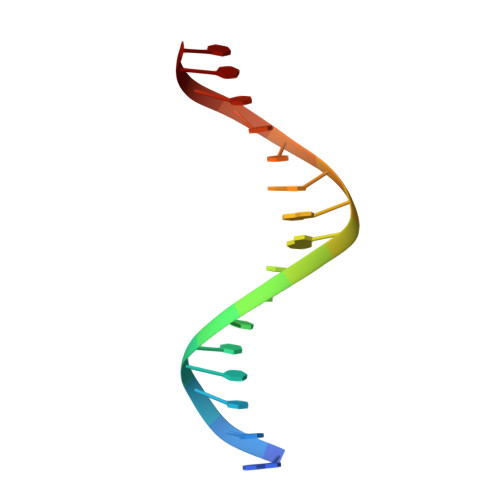
Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Xu, X., Li, Y., Bharath, S.R., Ozturk, M.B., Bowler, M.W., Loo, B.Z.L., Tergaonkar, V., Song, H.(2018) Nat Commun 9: 3183-3183
- PubMed: 30093619
- DOI: https://doi.org/10.1038/s41467-018-05644-0
- Primary Citation of Related Structures:
5ZMC - PubMed Abstract:
Transcriptional factors ETS1/2 and p52 synergize downstream of non-canonical NF-κB signaling to drive reactivation of the -146C>T mutant TERT promoter in multiple cancer types, but the mechanism underlying this cooperativity remains unknown. Here we report the crystal structure of a ternary p52/ETS1/-146C>T TERT promoter complex. While p52 needs to associate with consensus κB sites on the DNA to function during non-canonical NF-κB signaling, we show that p52 can activate the -146C>T TERT promoter without binding DNA. Instead, p52 interacts with ETS1 to form a heterotetramer, counteracting autoinhibition of ETS1. Analogous to observations with the GABPA/GABPB heterotetramer, the native flanking ETS motifs are required for sustained activation of the -146C>T TERT promoter by the p52/ETS1 heterotetramer. These observations provide a unifying mechanism for transcriptional activation by GABP and ETS1, and suggest that genome-wide targets of non-canonical NF-κB signaling are not limited to those driven by consensus κB sequences.
- Institute of Molecular and Cell Biology, 61 Biopolis Drive, Singapore, 138673, Singapore.
Organizational Affiliation: