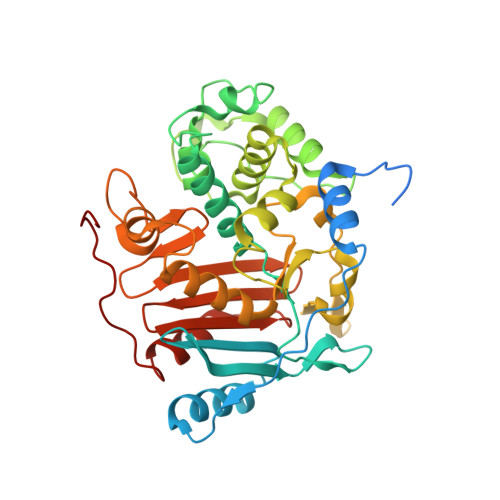
Repurposing an Ancient Protein Core Structure: Structural Studies on FmtA, a Novel Esterase of Staphylococcus aureus.
Dalal, V., Kumar, P., Rakhaminov, G., Qamar, A., Fan, X., Hunter, H., Tomar, S., Golemi-Kotra, D., Kumar, P.(2019) J Mol Biology 431: 3107-3123
- PubMed: 31260692
- DOI: https://doi.org/10.1016/j.jmb.2019.06.019
- Primary Citation of Related Structures:
5ZH8 - PubMed Abstract:
FmtA is a penicillin-recognizing protein (PRP) with novel hydrolytic activity toward the ester bond between d-Ala and the backbone of teichoic acids. Teichoic acids are polyol-phosphate polymers found in the Staphylococcus aureus cell wall, and they play important roles in antibiotic resistance and pathogenesis. Two of the PRPs conserved motifs, namely, SXXK and Y(S)XN, are involved in the hydrolysis by FmtA, but the catalytic mechanism remains elusive. Here we determined the crystal structure of FmtA. FmtA shares the core structure of PRPs: an all α-helical domain and α/β domain sandwiched together. However, it does not have the typical PRPs active-site cleft. Its active site is shallow, solvent-exposed, and enlarged. Furthermore, our mutagenesis and kinetic studies suggest that the SXXK and Y(S)XN motifs of FmtA offer all that is necessary for catalysis, and more: the active-site nucleophile (serine), the general base (lysine) required for the acylation step and the deacylation step, and an anchor (tyrosine) to hold the active-site serine, and possibly the substrate, in an optimum conformation for catalysis. Our study establishes that the FmtA esterase activity represents an expansion of the catalytic activity repertoire of the PRPs core structure, which typically displays peptidase activity. This finding points toward a novel mechanism of ester bond hydrolysis by a PRP. The structure of FmtA provides insights to the design of inhibitor molecules with the potential to serve as leads in the development of novel antibacterial chemotherapeutic agents.
- Department of Biotechnology, IIT, Roorkee, Uttrakhand-247667, India.
Organizational Affiliation: