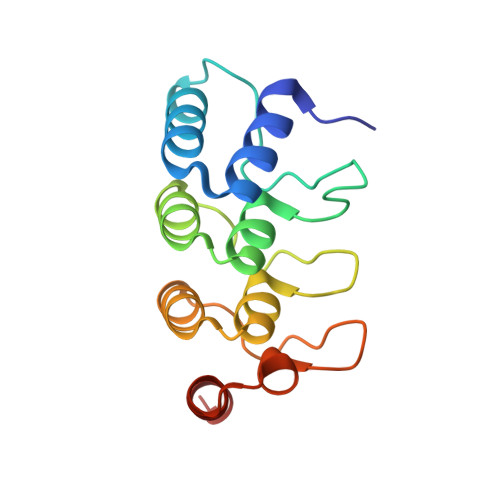
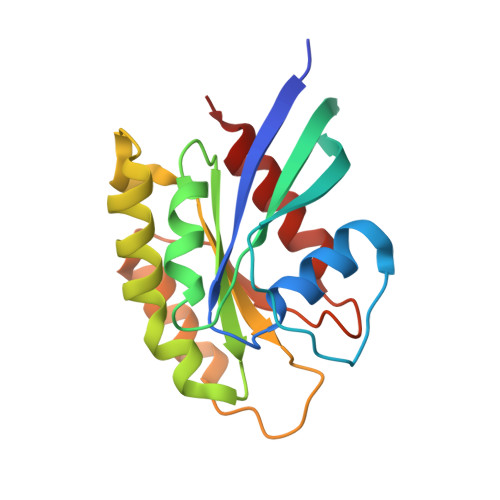
A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning.
Ma, X., Liu, K., Li, J., Li, H., Li, J., Liu, Y., Yang, C., Liang, H.(2018) J Biological Chem 293: 14155-14164
- PubMed: 30012887
- DOI: https://doi.org/10.1074/jbc.RA118.001854
- Primary Citation of Related Structures:
5Z2M, 5Z2N - PubMed Abstract:
Endosomal transport represents the primary mode for intracellular trafficking and signaling transduction and thus has to be tightly controlled. The molecular processes controlling the endosomal positioning utilize several large protein complexes, one of which contains the small GTPase Rab7, Rab-interacting lysosomal protein (RILP), and oxysterol-binding protein-related protein 1 (ORP1L). Rab7 is known to interact with RILP through a canonical binding site termed the effector-interacting switch region, but it is not clear how Rab7 interacts with ORP1L, limiting our understanding of the overall process. Here, we report structural and biochemical investigation of the Rab7-ORP1L interaction. We found that, contrary to prior studies, the interaction between Rab7 and the N-terminal ankyrin repeat domain (ARD N ) of ORP1L is independent of Rab7's GTP- or GDP-binding state. Moreover, we show that Rab7 interacts with ORP1L ARD N via a unique region consisting of helix3 (α3) and 3 10 -helix 2 (η2). This architecture leaves the canonical effector-interacting switch regions available for RILP binding and thus allows formation of the ORP1L-Rab7-RILP tripartite complex. Mutational disruption of the interacting interface between ORP1L and Rab7 compromised the ability of ORP1L-Rab7-RILP to regulate the late endosome positioning. Collectively, our results again manifested the versatility in the interaction between GTPase and its effector.
- From the National Laboratory of Biomacromolecules, Institute of Biophysics and.
Organizational Affiliation: