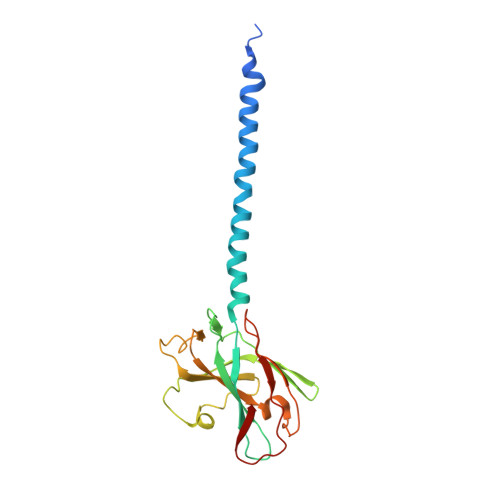
High-resolution archaellum structure reveals a conserved metal-binding site.
Meshcheryakov, V.A., Shibata, S., Schreiber, M.T., Villar-Briones, A., Jarrell, K.F., Aizawa, S.I., Wolf, M.(2019) EMBO Rep 20
- PubMed: 30898768
- DOI: https://doi.org/10.15252/embr.201846340
- Primary Citation of Related Structures:
5YA6, 5Z1L - PubMed Abstract:
Many archaea swim by means of archaella. While the archaellum is similar in function to its bacterial counterpart, its structure, composition, and evolution are fundamentally different. Archaella are related to archaeal and bacterial type IV pili. Despite recent advances, our understanding of molecular processes governing archaellum assembly and stability is still incomplete. Here, we determine the structures of Methanococcus archaella by X-ray crystallography and cryo-EM The crystal structure of Methanocaldococcus jannaschii FlaB1 is the first and only crystal structure of any archaellin to date at a resolution of 1.5 Å, which is put into biological context by a cryo-EM reconstruction from Methanococcus maripaludis archaella at 4 Å resolution created with helical single-particle analysis. Our results indicate that the archaellum is predominantly composed of FlaB1. We identify N-linked glycosylation by cryo-EM and mass spectrometry. The crystal structure reveals a highly conserved metal-binding site, which is validated by mass spectrometry and electron energy-loss spectroscopy. We show in vitro that the metal-binding site, which appears to be a widespread property of archaellin, is required for filament integrity.
- Molecular Cryo-Electron Microscopy Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Kunigami, Okinawa, Japan.
Organizational Affiliation: