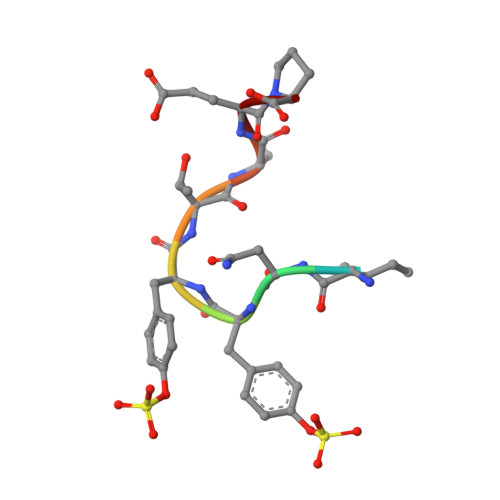
Tyrosine Sulfation Restricts the Conformational Ensemble of a Flexible Peptide, Strengthening the Binding Affinity for an Antibody
Miyanabe, K., Yamashita, T., Abe, Y., Akiba, H., Takamatsu, Y., Nakakido, M., Hamakubo, T., Ueda, T., Caaveiro, J.M.M., Tsumoto, K.(2018) Biochemistry 57: 4177-4185
- PubMed: 29936828
- DOI: https://doi.org/10.1021/acs.biochem.8b00592
- Primary Citation of Related Structures:
5YY4 - PubMed Abstract:
Protein tyrosine sulfation (PTS) is a post-translational modification regulating numerous biological events. PTS generally occurs at flexible regions of proteins, enhancing intermolecular interactions between proteins. Because of the high flexibility associated with the regions where PTS is generally encountered, an atomic-level understanding has been difficult to achieve by X-ray crystallography or nuclear magnetic resonance techniques. In this study, we focused on the conformational behavior of a flexible sulfated peptide and its interaction with an antibody. Molecular dynamics simulations and thermodynamic analysis indicated that PTS reduced the main-chain fluctuations upon the appearance of sulfate-mediated intramolecular H-bonds. Collectively, our data suggested that one of the mechanisms by which PTS may enhance protein-protein interactions consists of the limitation of conformational dynamics in the unbound state, thus reducing the loss of entropy upon binding and boosting the affinity for its partner.
- Department of Chemistry and Biotechnology, School of Engineering , The University of Tokyo , 7-3-1 Hongo , Bunkyo-ku, Tokyo 113-8656 , Japan.
Organizational Affiliation: