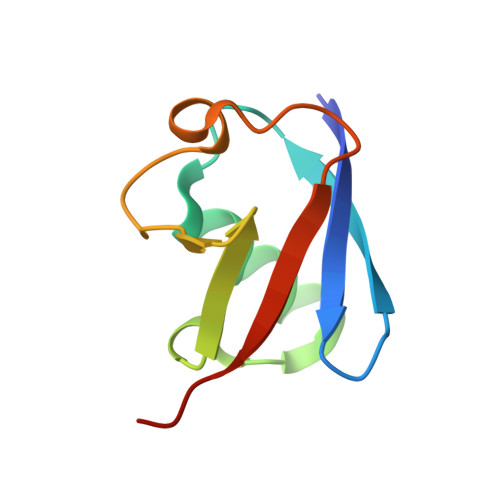
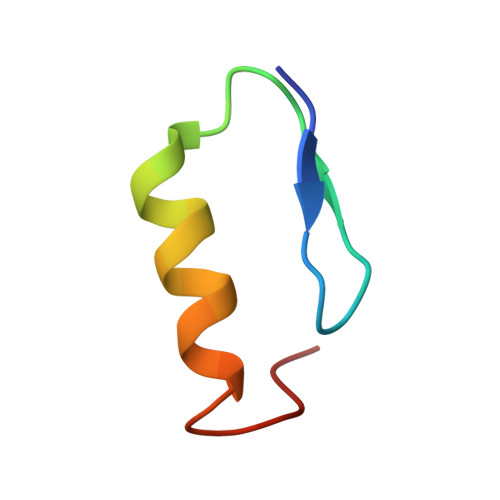
Mechanistic Insights into Recognitions of Ubiquitin and Myosin VI by Autophagy Receptor TAX1BP1.
Hu, S., Wang, Y., Gong, Y., Liu, J., Li, Y., Pan, L.(2018) J Mol Biology 430: 3283-3296
- PubMed: 29940186
- DOI: https://doi.org/10.1016/j.jmb.2018.06.030
- Primary Citation of Related Structures:
5YT6 - PubMed Abstract:
TAX1BP1, a ubiquitin-binding adaptor, plays critical roles in the innate immunity and selective autophagy. During autophagy, TAX1BP1 may not only function as an autophagy receptor to recruit ubiquitylated substrates for autophagic degradation, but also serve as a Myosin VI cargo adaptor protein for mediating the maturation of autophagosome. However, the mechanistic basis underlying the specific interactions of TAX1BP1 with ubiquitin and Myosin VI remains elusive. Here, using biochemical, NMR and structural analyses, we elucidate the detailed binding mechanism and uncover the key determinants for the interaction between TAX1BP1 and ubiquitin. In addition, we reveal that both tandem zinc-fingers of TAX1BP1 and the conformational rigidity between them are required for the Myosin VI binding of TAX1BP1, and ubiquitin and Myosin VI are mutually exclusive in binding to TAX1BP1. Collectively, our findings provide mechanistic insights into the dual functions of TAX1BP1 in selective autophagy.
- State Key Laboratory of Bioorganic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, 345 Lingling Road, Shanghai 200032, China.
Organizational Affiliation: