The Interaction between the Ribosomal Stalk Proteins and Translation Initiation Factor 5B Promotes Translation Initiation
Murakami, R., Singh, C.R., Morris, J., Tang, L., Harmon, I., Takasu, A., Miyoshi, T., Ito, K., Asano, K., Uchiumi, T.(2018) Mol Cell Biol 38
- PubMed: 29844065
- DOI: https://doi.org/10.1128/MCB.00067-18
- Primary Citation of Related Structures:
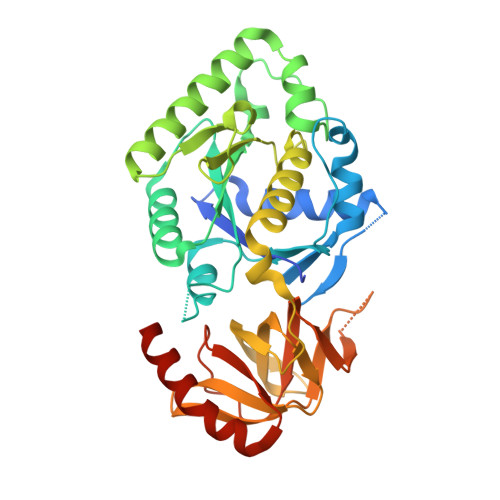
5YT0 - PubMed Abstract:
Ribosomal stalk proteins recruit translation elongation GTPases to the factor-binding center of the ribosome. Initiation factor 5B (eIF5B in eukaryotes and aIF5B in archaea) is a universally conserved GTPase that promotes the joining of the large and small ribosomal subunits during translation initiation. Here we show that aIF5B binds to the C-terminal tail of the stalk protein. In the cocrystal structure, the interaction occurs between the hydrophobic amino acids of the stalk C-terminal tail and a small hydrophobic pocket on the surface of the GTP-binding domain (domain I) of aIF5B. A substitution mutation altering the hydrophobic pocket of yeast eIF5B resulted in a marked reduction in ribosome-dependent eIF5B GTPase activity in vitro In yeast cells, the eIF5B mutation affected growth and impaired GCN4 expression during amino acid starvation via a defect in start site selection for the first upstream open reading frame in GCN4 mRNA, as observed with the eIF5B deletion mutant. The deletion of two of the four stalk proteins diminished polyribosome levels (indicating defective translation initiation) and starvation-induced GCN4 expression, both of which were suppressible by eIF5B overexpression. Thus, the mutual interaction between a/eIF5B and the ribosomal stalk plays an important role in subunit joining during translation initiation in vivo .
- Department of Biology, Faculty of Science, Niigata University, Niigata, Japan.
Organizational Affiliation: