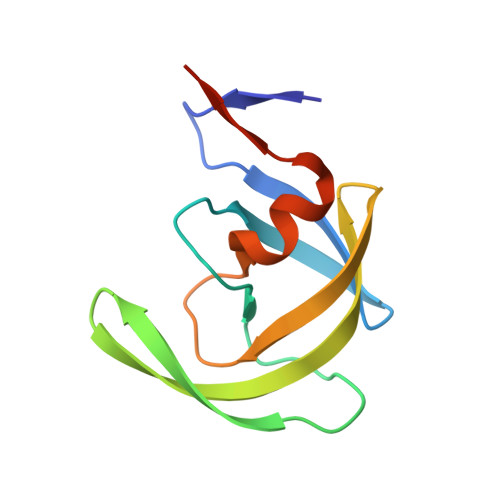
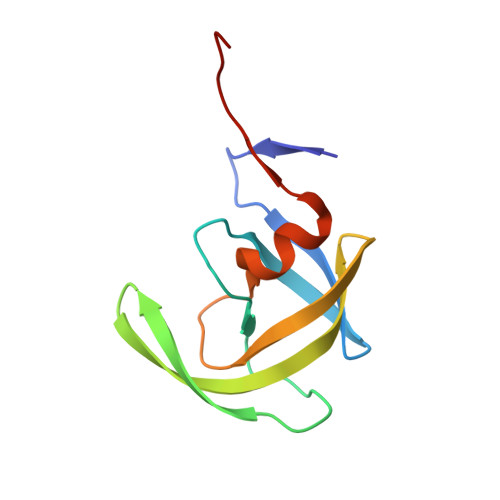
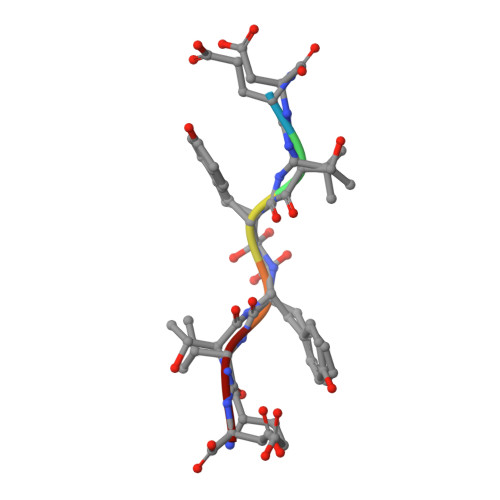
X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
Das, A., Mahale, S., Prashar, V., Bihani, S., Ferrer, J.L., Hosur, M.V.(2010) J Am Chem Soc 132: 6366-6373
- PubMed: 20397633
- DOI: https://doi.org/10.1021/ja100002b
- Primary Citation of Related Structures:
5YRS - PubMed Abstract:
Structural snapshots of each step in the catalytic cycle would help development of inhibitors of human immunodeficiency virus type 1 protease (HIV-1 PR) as effective drugs against HIV/AIDS. We report here one snapshot obtained by determining the structure of enzyme-substrate complex under conditions where the catalytic activity of the enzyme is greatly reduced. The 1.76 A crystal structure shows the oligopeptide substrate, AETFYVDGAA, converted in situ into a gem-diol tetrahedral intermediate (TI). The gem-diol intermediate is neutral and one of the hydroxyl oxygens forms a very short hydrogen bond (2.2 A) with the anionic aspartate of the catalytic dyad, which is monoprotonated. Further, there is no hydrogen atom on the outer oxygen of the neutral aspartate. These two observations provide direct evidence that, in the reaction mechanism, hydrogen bonding between catalytic aspartate and scissile carbonyl oxygen facilitates water attack on the scissile carbon atom. Comparison with the structural snapshot of the biproduct complex involving the same substrate reveals the reorganization of the hydrogen bonds at the catalytic center as the enzymatic reaction progresses toward completion. Accumulation of TI in the crystals provides direct evidence that collapse of TI is the rate-limiting step of hydrolysis.
- Protein Crystallography Section, Solid State Physics Division, Bhabha Atomic Research Centre, Trombay, Mumbai-400085, India.
Organizational Affiliation: