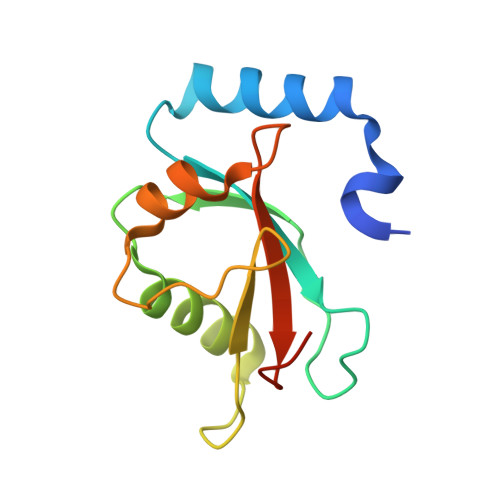
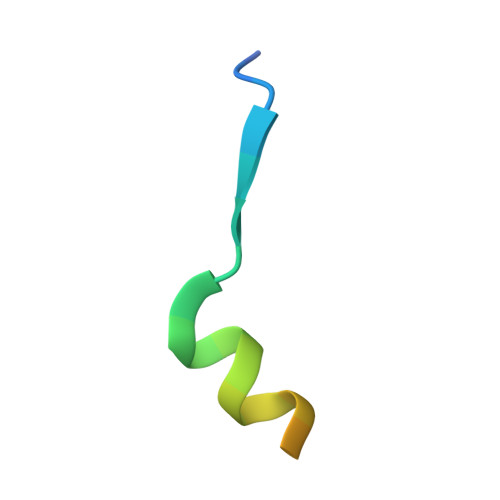
Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Li, J., Zhu, R., Chen, K., Zheng, H., Zhao, H., Yuan, C., Zhang, H., Wang, C., Zhang, M.(2018) Nat Chem Biol 14: 778-787
- PubMed: 29867141
- DOI: https://doi.org/10.1038/s41589-018-0082-8
- Primary Citation of Related Structures:
5YIP, 5YIQ, 5YIR, 5YIS - PubMed Abstract:
The mammalian Atg8 family proteins are central drivers of autophagy and contain six members, classified into the LC3 and GABARAP subfamilies. Due to their high sequence similarity and consequent functional overlaps, it is difficult to delineate specific functions of Atg8 proteins in autophagy. Here we discover a super-strong GABARAP-selective inhibitory peptide harbored in 270/480 kDa ankyrin-G and a super-potent pan-Atg8 inhibitory peptide from 440 kDa ankyrin-B. Structural studies elucidate the mechanism governing the Atg8 binding potency and selectivity of the peptides, reveal a general Atg8-binding sequence motif, and allow development of a more GABARAP-selective inhibitory peptide. These peptides effectively blocked autophagy when expressed in cultured cells. Expression of these ankyrin-derived peptides in Caenorhabditis elegans also inhibited autophagy, causing accumulation of the p62 homolog SQST-1, delayed development and shortened life span. Thus, these genetically encodable autophagy inhibitory peptides can be used to occlude autophagy spatiotemporally in living animals.
- Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China.
Organizational Affiliation: