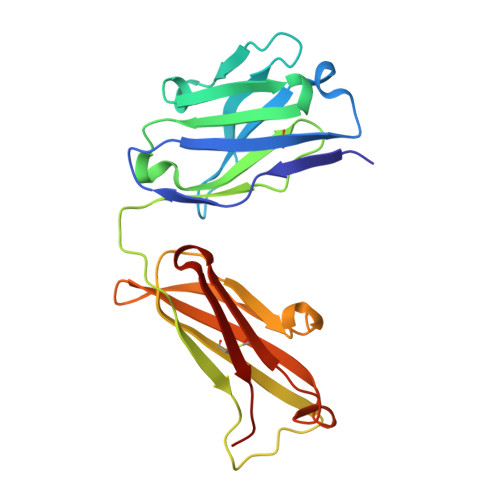
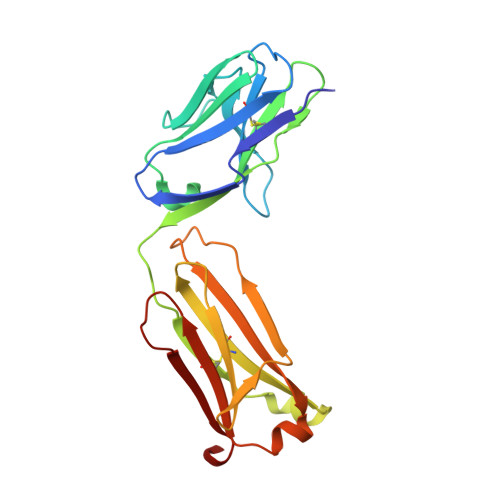
JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Handoko, L., Kaczkowski, B., Hon, C.C., Lizio, M., Wakamori, M., Matsuda, T., Ito, T., Jeyamohan, P., Sato, Y., Sakamoto, K., Yokoyama, S., Kimura, H., Minoda, A., Umehara, T.(2018) Epigenetics 13: 410-431
- PubMed: 30080437
- DOI: https://doi.org/10.1080/15592294.2018.1469891
- Primary Citation of Related Structures:
5YE3, 5YE4 - PubMed Abstract:
The bromodomain and extra-terminal domain (BET) proteins are promising drug targets for cancer and immune diseases. However, BET inhibition effects have been studied more in the context of bromodomain-containing protein 4 (BRD4) than BRD2, and the BET protein association to histone H4-hyperacetylated chromatin is not understood at the genome-wide level. Here, we report transcription start site (TSS)-resolution integrative analyses of ChIP-seq and transcriptome profiles in human non-small cell lung cancer (NSCLC) cell line H23. We show that di-acetylation at K5 and K8 of histone H4 (H4K5acK8ac) co-localizes with H3K27ac and BRD2 in the majority of active enhancers and promoters, where BRD2 has a stronger association with H4K5acK8ac than H3K27ac. Although BET inhibition by JQ1 led to complete reduction of BRD2 binding to chromatin, only local changes of H4K5acK8ac levels were observed, suggesting that recruitment of BRD2 does not influence global histone H4 hyperacetylation levels. This finding supports a model in which recruitment of BET proteins via histone H4 hyperacetylation is predominant over hyperacetylation of histone H4 by BET protein-associated acetyltransferases. In addition, we found that a remarkable number of BRD2-bound genes, including MYC and its downstream target genes, were transcriptionally upregulated upon JQ1 treatment. Using BRD2-enriched sites and transcriptional activity analysis, we identified candidate transcription factors potentially involved in the JQ1 response in BRD2-dependent and -independent manner.
- a Division of Genomic Technologies , RIKEN Center for Life Science Technologies, Yokohama , Kanagawa , Japan.
Organizational Affiliation: