Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Jang, J.Y., Bae, H., Lee, Y.J., Choi, Y.I., Kim, H.J., Park, S.B., Suh, S.W., Kim, S.W., Han, B.W.(2018) Sci Rep 8: 31-31
- PubMed: 29311579
- DOI: https://doi.org/10.1038/s41598-017-18274-1
- Primary Citation of Related Structures:
5YCN, 5YCP - PubMed Abstract:
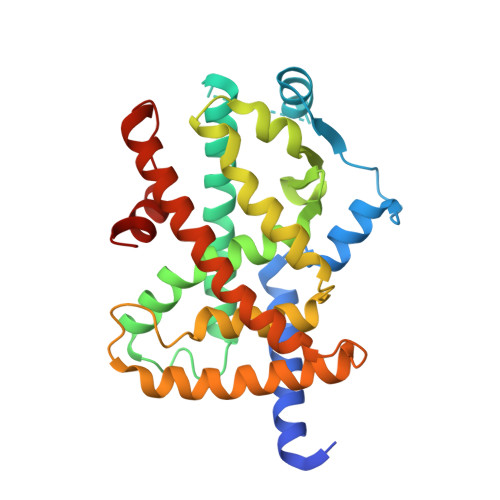
Peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear receptor superfamily. It functions as a ligand-activated transcription factor and plays important roles in the regulation of adipocyte differentiation, insulin resistance, and inflammation. Here, we report the crystal structures of PPARγ in complex with lobeglitazone, a novel PPARγ agonist, and with rosiglitazone for comparison. The thiazolidinedione (TZD) moiety of lobeglitazone occupies the canonical ligand-binding pocket near the activation function-2 (AF-2) helix (i.e., helix H12) in ligand-binding domain as the TZD moiety of rosiglitazone does. However, the elongated p-methoxyphenol moiety of lobeglitazone interacts with the hydrophobic pocket near the alternate binding site of PPARγ. The extended interaction of lobeglitazone with the hydrophobic pocket enhances its binding affinity and could affect the cyclin-dependent kinase 5 (Cdk5)-mediated phosphorylation of PPARγ at Ser245 (in PPARγ1 numbering; Ser273 in PPARγ2 numbering). Lobeglitazone inhibited the phosphorylation of PPARγ at Ser245 in a dose-dependent manner and exhibited a better inhibitory effect on Ser245 phosphorylation than rosiglitazone did. Our study provides new structural insights into the PPARγ regulation by TZD drugs and could be useful for the discovery of new PPARγ ligands as an anti-diabetic drug, minimizing known side effects.
- Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul, 08826, Republic of Korea.
Organizational Affiliation: