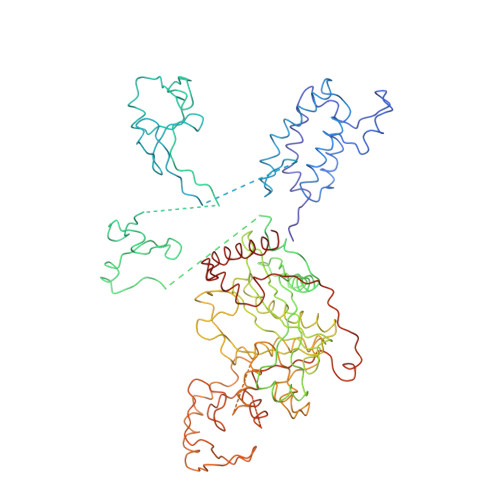
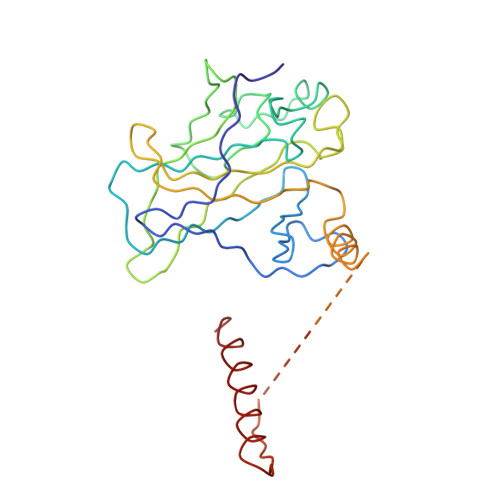
Tumor suppressor p53-mediated structural reorganization of the transcriptional coactivator p300.
Ghosh, R., Kaypee, S., Shasmal, M., Kundu, T.K., Roy, S., Sengupta, J.(2019) Biochemistry
- PubMed: 31314496
- DOI: https://doi.org/10.1021/acs.biochem.9b00333
- Primary Citation of Related Structures:
5XZC, 6K4N - PubMed Abstract:
Transcriptional coactivator p300, a critical player in eukaryotic gene regulation, primarily functions as a histone acetyltransferase (HAT). It is also an important player in acetylation of a number of nonhistone proteins, p53 being the most prominent one. Recruitment of p300 to p53 is pivotal in the regulation of p53-dependent genes. Emerging evidence suggests that p300 adopts an active conformation upon binding to the tetrameric p53, resulting in its enhanced acetylation activity. As a modular protein, p300 consists of multiple well-defined domains, where the structured domains are interlinked with unstructured linker regions. A crystal structure of the central domain of p300 encompassing Bromo, RING, PHD, and HAT domains demonstrates a compact module, where the HAT active site stays occluded by the RING domain. However, although p300 has a significant role in mediating the transcriptional activity of p53, only a few structural details on the complex of these two full-length proteins are available. Here, we present a cryo-electron microscopy (cryo-EM) study on the p300-p53 complex. The three-dimensional cryo-EM density map of the p300-p53 complex, when compared to the cryo-EM map of free p300, revealed that substantial change in the relative arrangement of Bromo and HAT domains occurs upon complex formation, which is likely required for exposing HAT active site and subsequent acetyltransferase activity. Our observation correlates well with previous studies showing that the presence of Bromodomain is obligatory for effective acetyltransferase activity of HAT. Thus, our result sheds new light on the mechanism whereby p300, following binding with p53, gets activated.
- Department of Biophysics , Bose Institute , Kolkata , India.
Organizational Affiliation: