Structure-activity studies of Mdm2/Mdm4-binding stapled peptides comprising non-natural amino acids.
Chee, S.M.Q., Wongsantichon, J., Siau, J., Thean, D., Ferrer, F., Robinson, R.C., Lane, D.P., Brown, C.J., Ghadessy, F.J.(2017) PLoS One 12: e0189379-e0189379
- PubMed: 29228061
- DOI: https://doi.org/10.1371/journal.pone.0189379
- Primary Citation of Related Structures:
5XXK - PubMed Abstract:
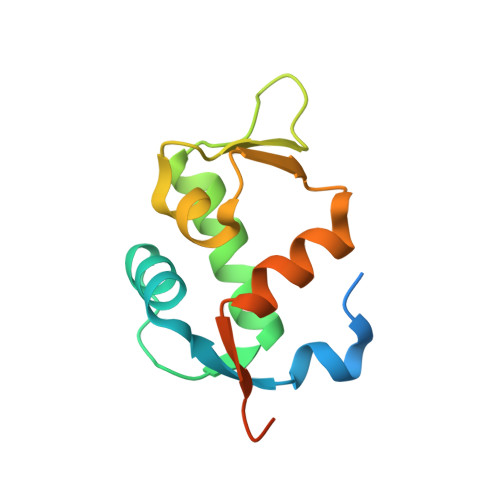
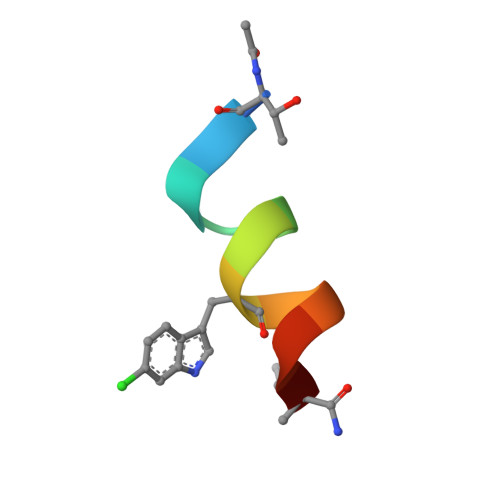
As primary p53 antagonists, Mdm2 and the closely related Mdm4 are relevant cancer therapeutic targets. We have previously described a series of cell-permeable stapled peptides that bind to Mdm2 with high affinity, resulting in activation of the p53 tumour suppressor. Within this series, highest affinity was obtained by modification of an obligate tryptophan residue to the non-natural L-6-chlorotryptophan. To understand the structural basis for improved affinity we have solved the crystal structure of this stapled peptide (M011) bound to Mdm2 (residues 6-125) at 1.66 Å resolution. Surprisingly, near identity to the structure of a related peptide (M06) without the 6-chloro modification is observed. Further analysis of linear and stapled peptides comprising 6-Me-tryptophan provides mechanistic insight into dual Mdm2/Mdm4 antagonism and confirms L98 of Mdm4 as a mutable steric gate. The results also highlight a possible role of the flexible hinge region in determining Mdm2/Mdm4 plasticity.
- p53Lab, Agency for Science Technology and Research (A*STAR), Singapore, Singapore.
Organizational Affiliation: