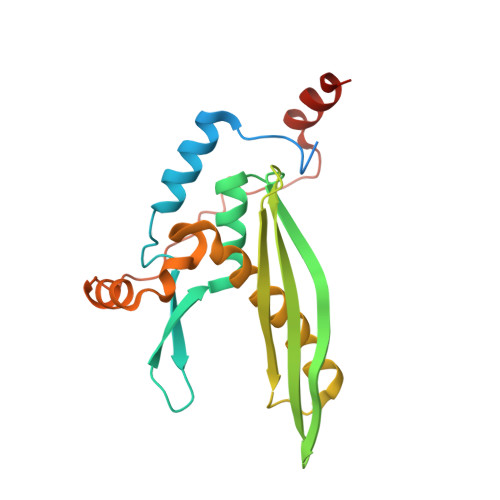
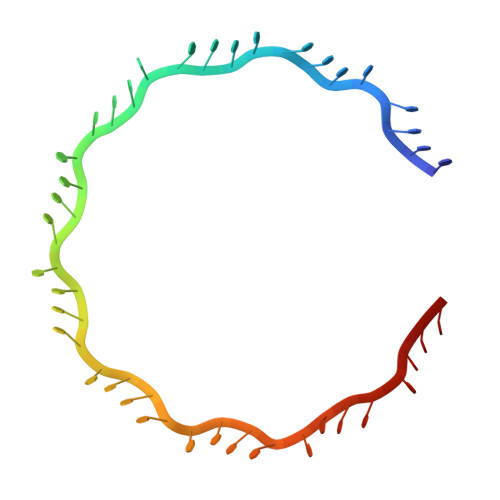
Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
Saotome, M., Saito, K., Yasuda, T., Ohtomo, H., Sugiyama, S., Nishimura, Y., Kurumizaka, H., Kagawa, W.(2018) iScience 3: 50-62
- PubMed: 30428330
- DOI: https://doi.org/10.1016/j.isci.2018.04.005
- Primary Citation of Related Structures:
5XRZ, 5XS0 - PubMed Abstract:
RAD52 mediates homologous recombination by annealing cDNA strands. However, the detailed mechanism of DNA annealing promoted by RAD52 has remained elusive. Here we report two crystal structures of human RAD52 single-stranded DNA (ssDNA) complexes that probably represent key reaction intermediates of RAD52-mediated DNA annealing. The first structure revealed a "wrapped" conformation of ssDNA around the homo-oligomeric RAD52 ring, in which the edges of the bases involved in base pairing are exposed to the solvent. The ssDNA conformation is close to B-form and appears capable of engaging in Watson-Crick base pairing with the cDNA strand. The second structure revealed a "trapped" conformation of ssDNA between two RAD52 rings. This conformation is stabilized by a different RAD52 DNA binding site, which promotes the accumulation of multiple RAD52 rings on ssDNA and the aggregation of ssDNA. These structures provide a structural framework for understanding the mechanism of RAD52-mediated DNA annealing.
- Department of Chemistry, Graduate School of Science and Engineering, Meisei University, 2-1-1 Hodokubo, Hino-shi, Tokyo 191-8506, Japan.
Organizational Affiliation: