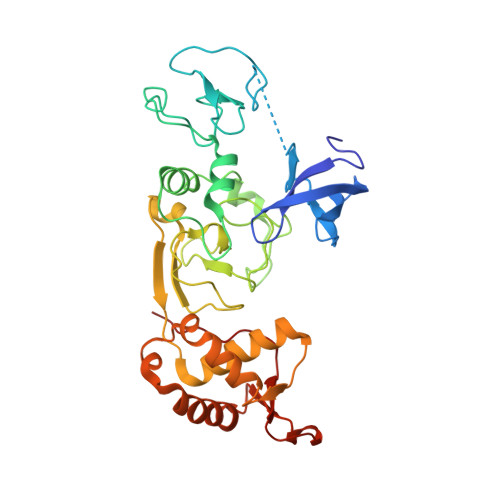
Polycomb-like proteins link the PRC2 complex to CpG islands
Li, H., Liefke, R., Jiang, J., Kurland, J.V., Tian, W., Deng, P., Zhang, W., He, Q., Patel, D.J., Bulyk, M.L., Shi, Y., Wang, Z.(2017) Nature 549: 287-291
- PubMed: 28869966
- DOI: https://doi.org/10.1038/nature23881
- Primary Citation of Related Structures:
5XFN, 5XFO, 5XFP, 5XFQ, 5XFR - PubMed Abstract:
The Polycomb repressive complex 2 (PRC2) mainly mediates transcriptional repression and has essential roles in various biological processes including the maintenance of cell identity and proper differentiation. Polycomb-like (PCL) proteins, such as PHF1, MTF2 and PHF19, are PRC2-associated factors that form sub-complexes with PRC2 core components, and have been proposed to modulate the enzymatic activity of PRC2 or the recruitment of PRC2 to specific genomic loci. Mammalian PRC2-binding sites are enriched in CG content, which correlates with CpG islands that display a low level of DNA methylation. However, the mechanism of PRC2 recruitment to CpG islands is not fully understood. Here we solve the crystal structures of the N-terminal domains of PHF1 and MTF2 with bound CpG-containing DNAs in the presence of H3K36me3-containing histone peptides. We show that the extended homologous regions of both proteins fold into a winged-helix structure, which specifically binds to the unmethylated CpG motif but in a completely different manner from the canonical winged-helix DNA recognition motif. We also show that the PCL extended homologous domains are required for efficient recruitment of PRC2 to CpG island-containing promoters in mouse embryonic stem cells. Our research provides the first, to our knowledge, direct evidence to demonstrate that PCL proteins are crucial for PRC2 recruitment to CpG islands, and further clarifies the roles of these proteins in transcriptional regulation in vivo.
- Key Laboratory of Cell Proliferation and Regulation Biology of Ministry of Education, College of Life Sciences, Beijing Normal University, 19 Xinjiekouwai Avenue, Beijing 100875, China.
Organizational Affiliation: