Structural basis for tRNA-dependent cysteine biosynthesis
Chen, M., Kato, K., Kubo, Y., Tanaka, Y., Liu, Y., Long, F., Whitman, W.B., Lill, P., Gatsogiannis, C., Raunser, S., Shimizu, N., Shinoda, A., Nakamura, A., Tanaka, I., Yao, M.(2017) Nat Commun 8: 1521-1521
- PubMed: 29142195
- DOI: https://doi.org/10.1038/s41467-017-01543-y
- Primary Citation of Related Structures:
5X6B, 5X6C - PubMed Abstract:
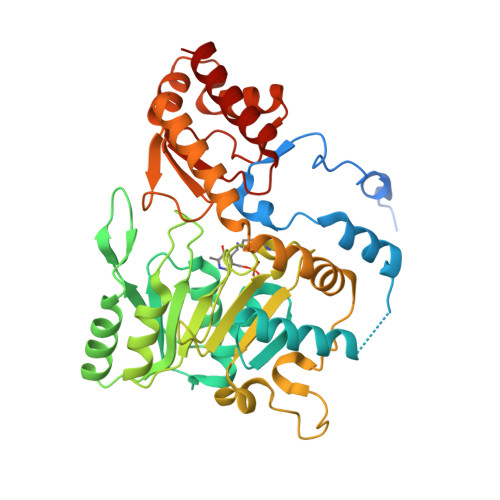
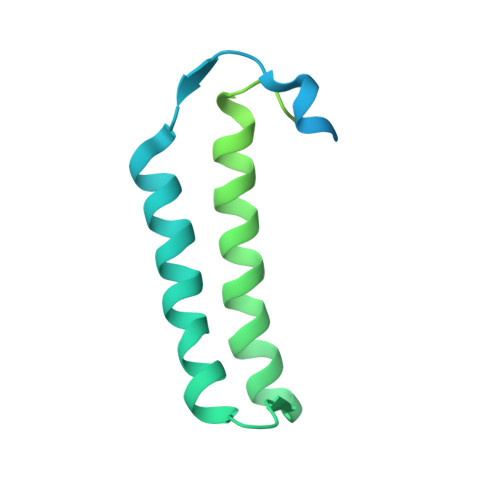
Cysteine can be synthesized by tRNA-dependent mechanism using a two-step indirect pathway, where O-phosphoseryl-tRNA synthetase (SepRS) catalyzes the ligation of a mismatching O-phosphoserine (Sep) to tRNA Cys followed by the conversion of tRNA-bounded Sep into cysteine by Sep-tRNA:Cys-tRNA synthase (SepCysS). In ancestral methanogens, a third protein SepCysE forms a bridge between the two enzymes to create a ternary complex named the transsulfursome. By combination of X-ray crystallography, SAXS and EM, together with biochemical evidences, here we show that the three domains of SepCysE each bind SepRS, SepCysS, and tRNA Cys , respectively, which mediates the dynamic architecture of the transsulfursome and thus enables a global long-range channeling of tRNA Cys between SepRS and SepCysS distant active sites. This channeling mechanism could facilitate the consecutive reactions of the two-step indirect pathway of Cys-tRNA Cys synthesis (tRNA-dependent cysteine biosynthesis) to prevent challenge of translational fidelity, and may reflect the mechanism that cysteine was originally added into genetic code.
- Graduate School of Life Science, Hokkaido University, Sapporo, 060-0810, Japan.
Organizational Affiliation: