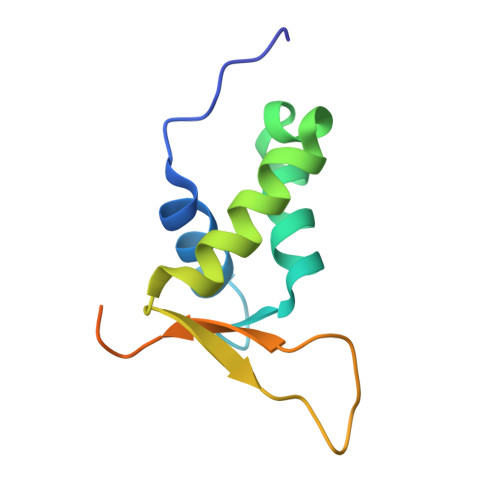
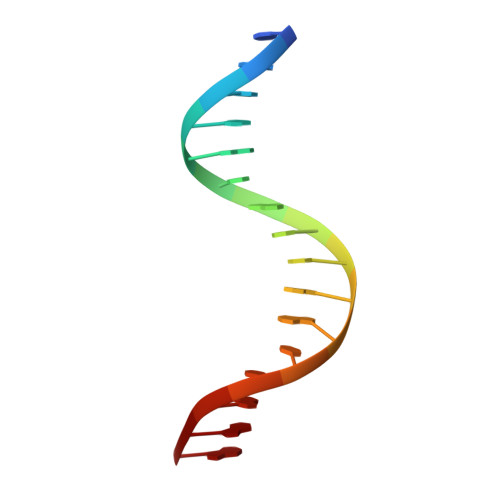
Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Li, J., Machado, A.C.D., Guo, M., Sagendorf, J.M., Zhou, Z., Jiang, L., Chen, X., Wu, D., Qu, L., Chen, Z., Chen, L., Rohs, R., Chen, Y.(2017) Biochemistry 56: 3745-3753
- PubMed: 28644006
- DOI: https://doi.org/10.1021/acs.biochem.7b00211
- Primary Citation of Related Structures:
5X07 - PubMed Abstract:
FOXA2, a member of the forkhead family of transcription factors, plays essential roles in liver development and bile acid homeostasis. In this study, we report a 2.8 Å co-crystal structure of the FOXA2 DNA-binding domain (FOXA2-DBD) bound to a DNA duplex containing a forkhead consensus binding site (GTAAACA). The FOXA2-DBD adopts the canonical winged-helix fold, with helix H3 and wing 1 regions mainly mediating the DNA recognition. Although the wing 2 region was not defined in the structure, isothermal titration calorimetry assays suggested that this region was required for optimal DNA binding. Structure comparison with the FOXA3-DBD bound to DNA revealed more major groove contacts and fewer minor groove contacts in the FOXA2 structure than in the FOXA3 structure. Structure comparison with the FOXO1-DBD bound to DNA showed that different forkhead proteins could induce different DNA conformations upon binding to identical DNA sequences. Our findings provide the structural basis for FOXA2 protein binding to a consensus forkhead site and elucidate how members of the forkhead protein family bind different DNA sites.
- Key Laboratory of Cancer Proteomics of Chinese Ministry of Health and Laboratory of Structural Biology, Xiangya Hospital, Central South University , Changsha, Hunan 410008, China.
Organizational Affiliation: