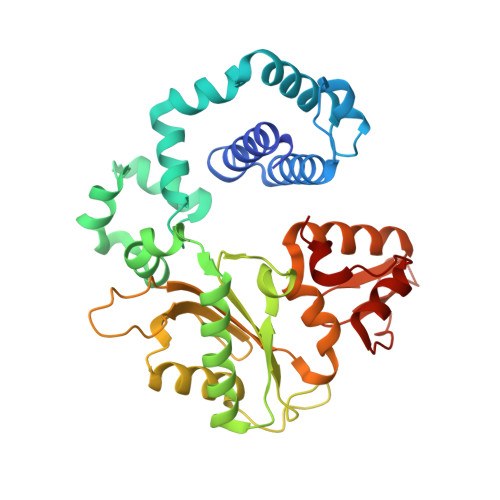
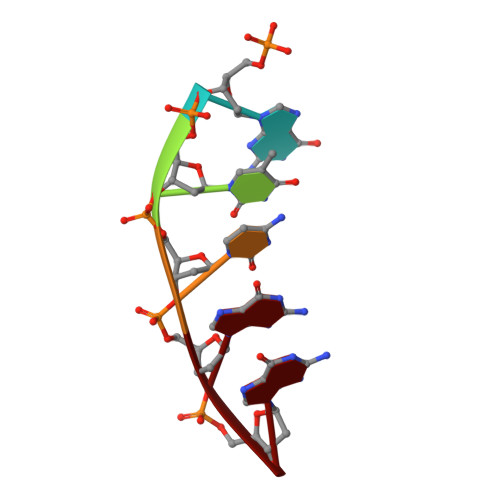
Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Schaich, M.A., Smith, M.R., Cloud, A.S., Holloran, S.M., Freudenthal, B.D.(2017) Chem Res Toxicol 30: 1993-2001
- PubMed: 28862449
- DOI: https://doi.org/10.1021/acs.chemrestox.7b00173
- Primary Citation of Related Structures:
5WNX, 5WNY, 5WNZ, 5WO0 - PubMed Abstract:
Members of the nucleoside analogue class of cancer therapeutics compete with canonical nucleotides to disrupt numerous cellular processes, including nucleotide homeostasis, DNA and RNA synthesis, and nucleotide metabolism. Nucleoside analogues are triphosphorylated and subsequently inserted into genomic DNA, contributing to the efficacy of therapeutic nucleosides in multiple ways. In some cases, the altered base acts as a mutagen, altering the DNA sequence to promote cellular death; in others, insertion of the altered nucleotide triggers DNA repair pathways, which produce lethal levels of cytotoxic intermediates such as single and double stranded DNA breaks. As a prerequisite to many of these biological outcomes, the modified nucleotide must be accommodated in the DNA polymerase active site during nucleotide insertion. Currently, the molecular contacts that mediate DNA polymerase insertion of modified nucleotides remain unknown for multiple therapeutic compounds, despite decades of clinical use. To determine how modified bases are inserted into duplex DNA, we used mammalian DNA polymerase β (pol β) to visualize the structural conformations of four therapeutically relevant modified nucleotides, 6-thio-2'-deoxyguanosine-5'-triphosphate (6-TdGTP), 5-fluoro-2'-deoxyuridine-5'-triphosphate (5-FdUTP), 5-formyl-deoxycytosine-5'-triphosphate (5-FodCTP), and 5-formyl-deoxyuridine-5'-triphosphate (5-FodUTP). Together, the structures reveal a pattern in which the modified nucleotides utilize Watson-Crick base pairing interactions similar to that of unmodified nucleotides. The nucleotide modifications were consistently positioned in the major groove of duplex DNA, accommodated by an open cavity in pol β. These results provide novel information for the rational design of new therapeutic nucleoside analogues and a greater understanding of how modified nucleotides are tolerated by polymerases.
- Department of Biochemistry and Molecular Biology, University of Kansas Medical Center , Kansas City, Kansas 66160, United States.
Organizational Affiliation: