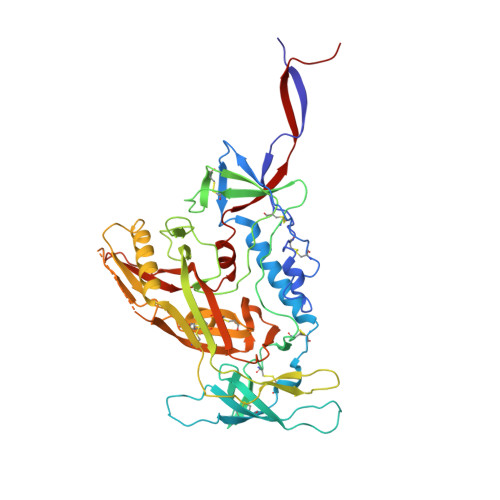
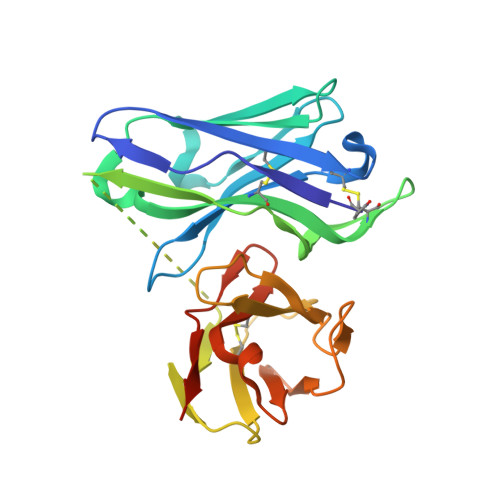
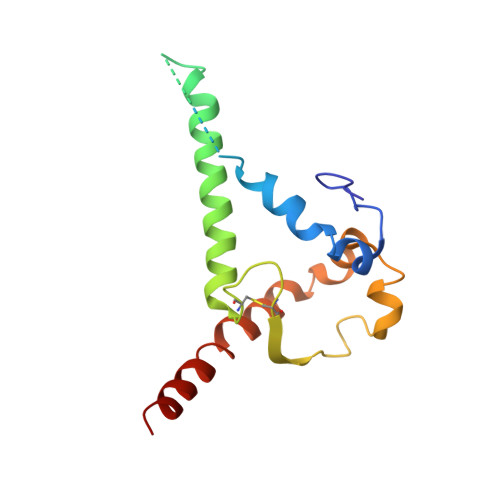
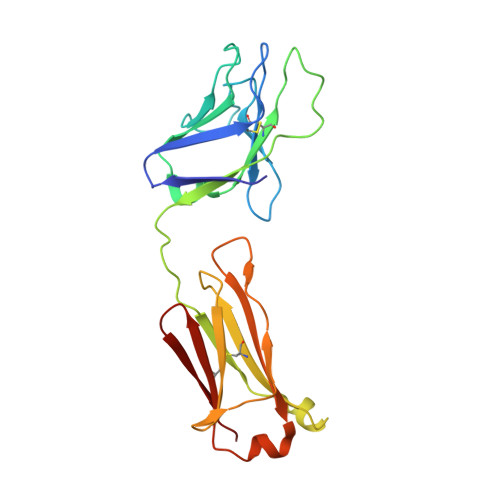
Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization.
Torrents de la Pena, A., Julien, J.P., de Taeye, S.W., Garces, F., Guttman, M., Ozorowski, G., Pritchard, L.K., Behrens, A.J., Go, E.P., Burger, J.A., Schermer, E.E., Sliepen, K., Ketas, T.J., Pugach, P., Yasmeen, A., Cottrell, C.A., Torres, J.L., Vavourakis, C.D., van Gils, M.J., LaBranche, C., Montefiori, D.C., Desaire, H., Crispin, M., Klasse, P.J., Lee, K.K., Moore, J.P., Ward, A.B., Wilson, I.A., Sanders, R.W.(2017) Cell Rep 20: 1805-1817
- PubMed: 28834745
- DOI: https://doi.org/10.1016/j.celrep.2017.07.077
- Primary Citation of Related Structures:
5WDU - PubMed Abstract:
The production of native-like recombinant versions of the HIV-1 envelope glycoprotein (Env) trimer requires overcoming the natural flexibility and instability of the complex. The engineered BG505 SOSIP.664 trimer mimics the structure and antigenicity of native Env. Here, we describe how the introduction of new disulfide bonds between the glycoprotein (gp)120 and gp41 subunits of SOSIP trimers of the BG505 and other genotypes improves their stability and antigenicity, reduces their conformational flexibility, and helps maintain them in the unliganded conformation. The resulting next-generation SOSIP.v5 trimers induce strong autologous tier-2 neutralizing antibody (NAb) responses in rabbits. In addition, the BG505 SOSIP.v6 trimers induced weak heterologous NAb responses against a subset of tier-2 viruses that were not elicited by the prototype BG505 SOSIP.664. These stabilization methods can be applied to trimers from multiple genotypes as components of multivalent vaccines aimed at inducing broadly NAbs (bNAbs).
- Department of Medical Microbiology, Academic Medical Center, University of Amsterdam, Amsterdam 1105 AZ, the Netherlands.
Organizational Affiliation: