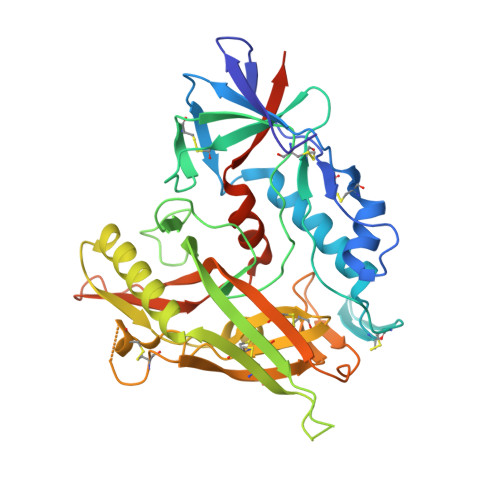
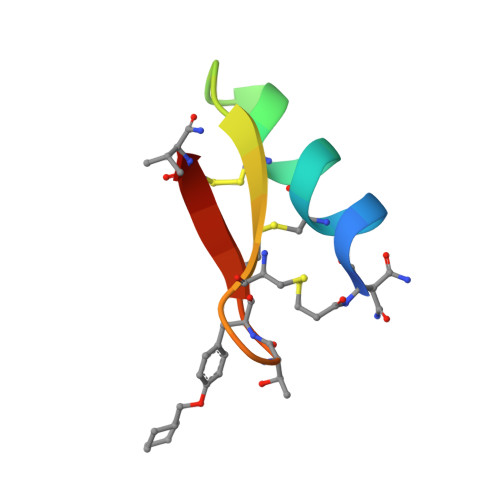
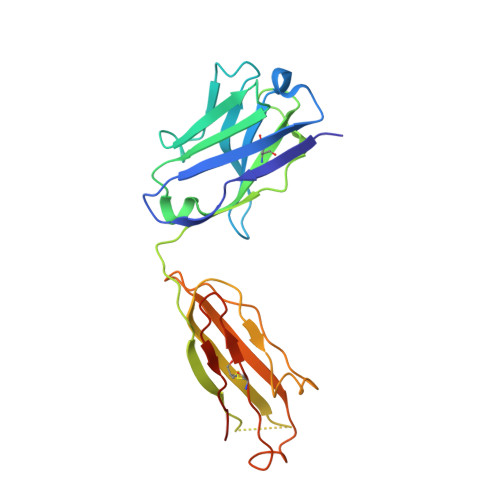
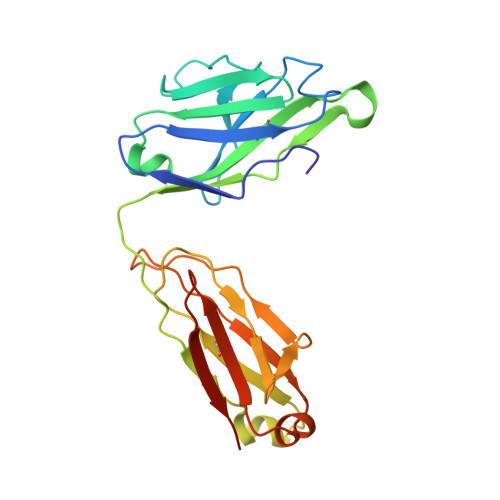
Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Tolbert, W.D., Gohain, N., Alsahafi, N., Van, V., Orlandi, C., Ding, S., Martin, L., Finzi, A., Lewis, G.K., Ray, K., Pazgier, M.(2017) Structure 25: 1719-1731.e4
- PubMed: 29056481
- DOI: https://doi.org/10.1016/j.str.2017.09.009
- Primary Citation of Related Structures:
5W4L - PubMed Abstract:
Antibodies can have an impact on HIV-1 infection in multiple ways, including antibody-dependent cellular cytotoxicity (ADCC), a correlate of protection observed in the RV144 vaccine trial. One of the most potent ADCC-inducing epitopes on HIV-1 Env is recognized by the C11 antibody. Here, we present the crystal structure, at 2.9 Å resolution, of the C11-like antibody N12-i3, in a quaternary complex with the HIV-1 gp120, a CD4-mimicking peptide M48U1, and an A32-like antibody, N5-i5. Antibody N12-i3 recognizes an epitope centered on the N-terminal "eighth strand" of a critical β sandwich, which our analysis indicates to be emblematic of a late-entry state, after the gp120 detachment. In prior entry states, this sandwich comprises only seven strands, with the eighth strand instead pairing with a portion of the gp120 C terminus. The conformational gymnastics of HIV-1 gp120 thus includes altered β-strand pairing, possibly to reduce immunogenicity, although nevertheless still recognized by the human immune system.
- Division of Vaccine Research, Institute of Human Virology, Baltimore, MD, USA; Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 725 West Lombard Street, Baltimore, MD 21201, USA.
Organizational Affiliation: