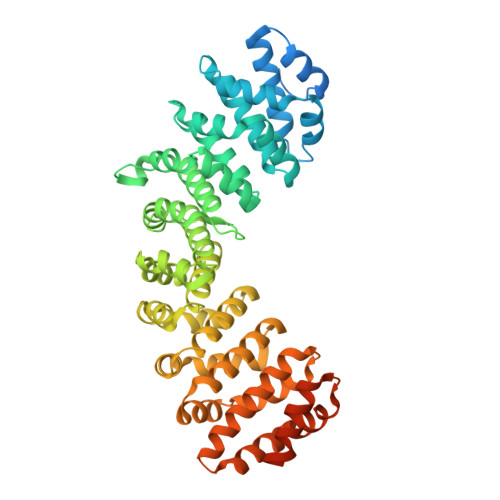
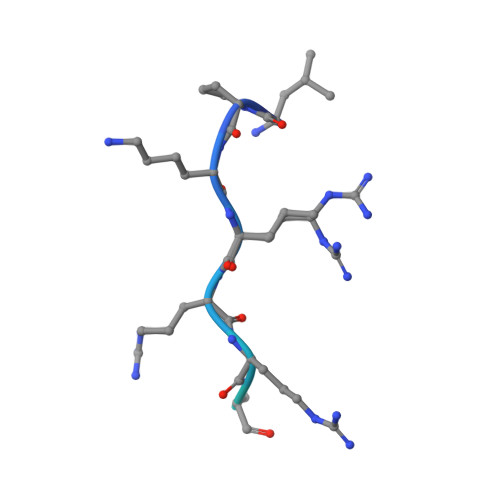
Variations in nuclear localization strategies among pol X family enzymes.
Kirby, T.W., Pedersen, L.C., Gabel, S.A., Gassman, N.R., London, R.E.(2018) Traffic
- PubMed: 29931796
- DOI: https://doi.org/10.1111/tra.12600
- Primary Citation of Related Structures:
5W4E, 5W4F, 6D7M, 6D7N - PubMed Abstract:
Despite the essential roles of pol X family enzymes in DNA repair, information about the structural basis of their nuclear import is limited. Recent studies revealed the unexpected presence of a functional nuclear localization signal (NLS) in DNA polymerase β, indicating the importance of active nuclear targeting, even for enzymes likely to leak into and out of the nucleus. The current studies further explore the active nuclear transport of these enzymes by identifying and structurally characterizing the functional NLS sequences in the three remaining human pol X enzymes: terminal deoxynucleotidyl transferase (TdT), DNA polymerase mu (pol μ) and DNA polymerase lambda (pol λ). NLS identifications are based on Importin α (Impα) binding affinity determined by fluorescence polarization of fluorescein-labeled NLS peptides, X-ray crystallographic analysis of the Impα∆IBB•NLS complexes and fluorescence-based subcellular localization studies. All three polymerases use NLS sequences located near their N-terminus; TdT and pol μ utilize monopartite NLS sequences, while pol λ utilizes a bipartite sequence, unique among the pol X family members. The pol μ NLS has relatively weak measured affinity for Impα, due in part to its proximity to the N-terminus that limits non-specific interactions of flanking residues preceding the NLS. However, this effect is partially mitigated by an N-terminal sequence unsupportive of Met1 removal by methionine aminopeptidase, leading to a 3-fold increase in affinity when the N-terminal methionine is present. Nuclear targeting is unique to each pol X family enzyme with variations dependent on the structure and unique functional role of each polymerase.
- National Institute of Environmental Health Sciences, Genome Integrity and Structural Biology Laboratory, National Institutes of Health, Research Triangle Park, North Carolina.
Organizational Affiliation: