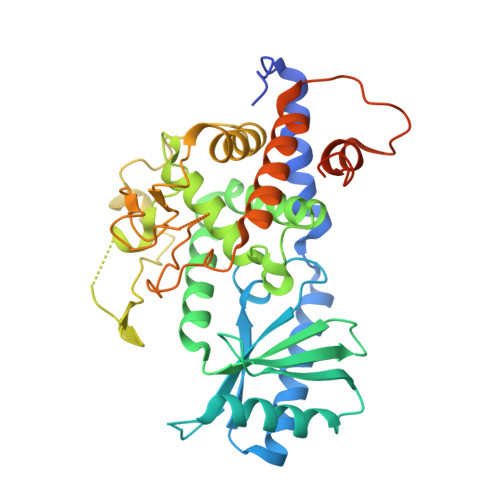
Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Faehnle, C.R., Walleshauser, J., Joshua-Tor, L.(2017) Nat Struct Mol Biol 24: 658-665
- PubMed: 28671666
- DOI: https://doi.org/10.1038/nsmb.3428
- Primary Citation of Related Structures:
5W0B, 5W0M, 5W0N, 5W0O - PubMed Abstract:
The uridyl transferases TUT4 and TUT7 (collectively called TUT4(7)) switch between two modes of activity, either promoting expression of let-7 microRNA (monoU) or marking it for degradation (oligoU). Lin28 modulates the switch via recruitment of TUT4(7) to the precursor pre-let-7 in stem cells and human cancers. We found that TUT4(7) utilize two multidomain functional modules during the switch from monoU to oligoU. The catalytic module (CM) is essential for both activities, while the Lin28-interacting module (LIM) is indispensable for oligoU. A TUT7 CM structure trapped in the monoU activity staterevealed a duplex-RNA-binding pocket that orients group II pre-let-7 hairpins to favor monoU addition. Conversely, the switch to oligoU requires the ZK domain of Lin28 to drive the formation of a stable ternary complex between pre-let-7 and the inactive LIM. Finally, ZK2 of TUT4(7) aids oligoU addition by engaging the growing oligoU tail through uracil-specific interactions.
- W.M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Organizational Affiliation: