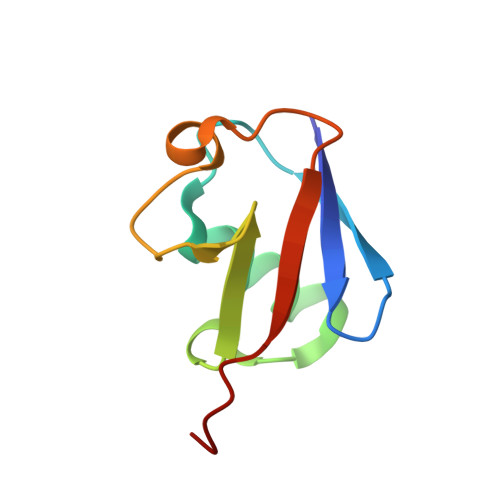
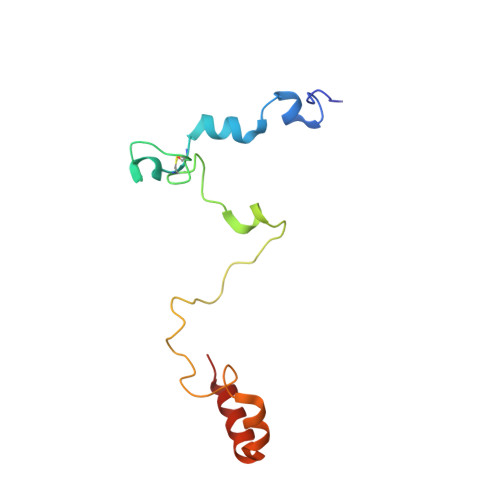
Structural Basis for the Interaction of Mutasome Assembly Factor REV1 with Ubiquitin.
Cui, G., Botuyan, M.V., Mer, G.(2018) J Mol Biology 430: 2042-2050
- PubMed: 29778604
- DOI: https://doi.org/10.1016/j.jmb.2018.05.017
- Primary Citation of Related Structures:
5VZM - PubMed Abstract:
REV1 is an evolutionarily conserved translesion synthesis (TLS) DNA polymerase and an assembly factor key for the recruitment of other TLS polymerases to DNA damage sites. REV1-mediated recognition of ubiquitin in the proliferative cell nuclear antigen is thought to be the trigger for TLS activation. Here we report the solution NMR structure of a 108-residue fragment of human REV1 encompassing the two putative ubiquitin-binding motifs UBM1 and UBM2 in complex with ubiquitin. While in mammals UBM1 and UBM2 are both required for optimal association of REV1 with replication factories after DNA damage, we show that only REV1 UBM2 binds ubiquitin. Structure-guided mutagenesis in Saccharomyces cerevisiae further highlights the importance of UBM2 for REV1-mediated mutagenesis and DNA damage tolerance.
- Department of Biochemistry and Molecular Biology, Mayo Clinic, 200 First Street SW, Rochester, MN 55905, USA.
Organizational Affiliation: