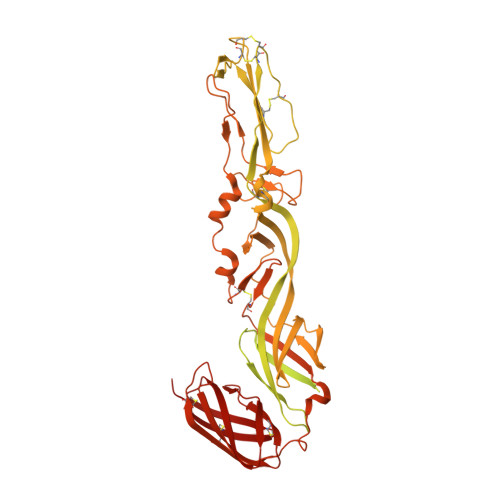
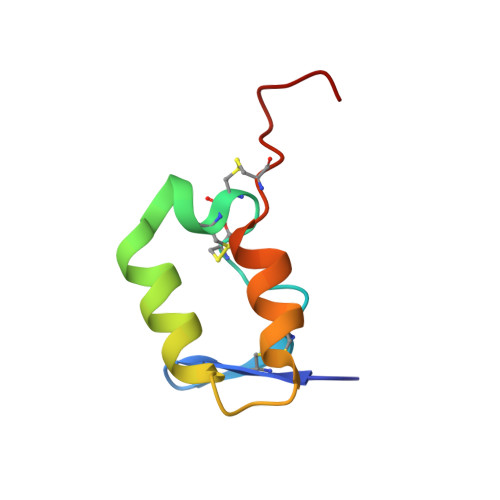
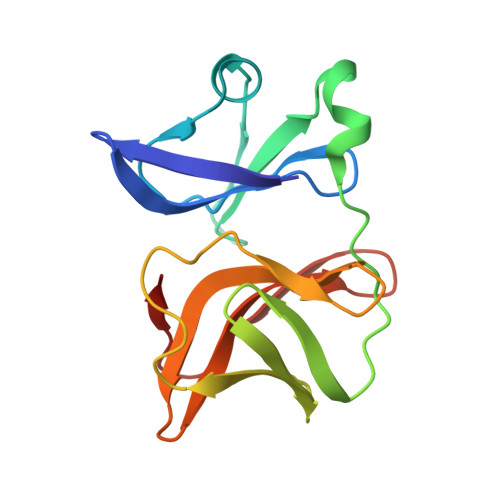
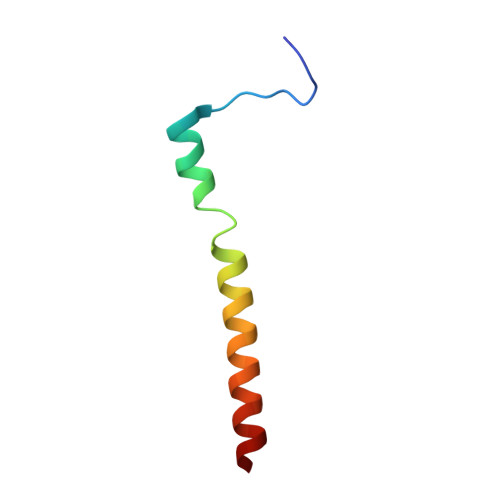
Structural studies of Chikungunya virus maturation.
Yap, M.L., Klose, T., Urakami, A., Hasan, S.S., Akahata, W., Rossmann, M.G.(2017) Proc Natl Acad Sci U S A 114: 13703-13707
- PubMed: 29203665
- DOI: https://doi.org/10.1073/pnas.1713166114
- Primary Citation of Related Structures:
5VU2 - PubMed Abstract:
Cleavage of the alphavirus precursor glycoprotein p62 into the E2 and E3 glycoproteins before assembly with the nucleocapsid is the key to producing fusion-competent mature spikes on alphaviruses. Here we present a cryo-EM, 6.8-Å resolution structure of an "immature" Chikungunya virus in which the cleavage site has been mutated to inhibit proteolysis. The spikes in the immature virus have a larger radius and are less compact than in the mature virus. Furthermore, domains B on the E2 glycoproteins have less freedom of movement in the immature virus, keeping the fusion loops protected under domain B. In addition, the nucleocapsid of the immature virus is more compact than in the mature virus, protecting a conserved ribosome-binding site in the capsid protein from exposure. These differences suggest that the posttranslational processing of the spikes and nucleocapsid is necessary to produce infectious virus.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.
Organizational Affiliation: