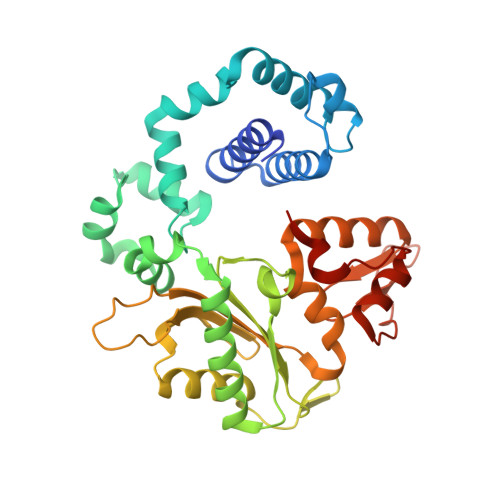
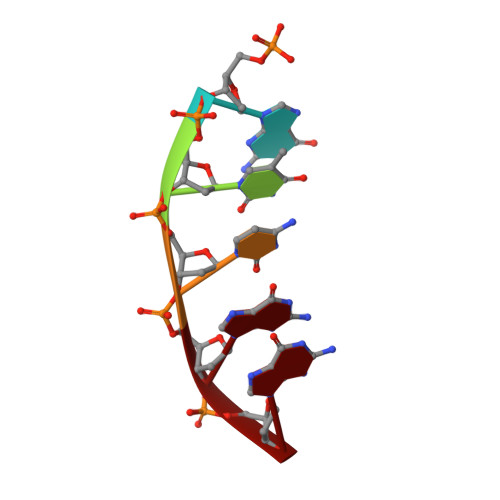
Time-Dependent Extension from an 8-Oxoguanine Lesion by Human DNA Polymerase Beta.
Reed, A.J., Suo, Z.(2017) J Am Chem Soc 139: 9684-9690
- PubMed: 28682600
- DOI: https://doi.org/10.1021/jacs.7b05048
- Primary Citation of Related Structures:
5VRW, 5VRX, 5VRY, 5VRZ, 5VS0, 5VS1, 5VS2, 5VS3, 5VS4 - PubMed Abstract:
The oxidative DNA lesion 7,8-dihydro-2'-deoxyguanine (8-oxoG) often occurs in double-stranded DNA and poses a threat to genomic integrity due to the ability of 8-oxoG to form stable Watson-Crick base pairs with deoxycytidine (8-oxoG:dC) and Hoogsteen base pairs with deoxyadenosine (8-oxoG:dA). In humans, short-patch base excision repair of 8-oxoG:dA base pairs requires human DNA polymerase β (hPolβ) to bypass 8-oxoG. Previously, we have shown hPolβ-catalyzed 8-oxoG bypass to exhibit low fidelity and identified a unique stacking interaction between the newly incorporated nucleotide (dCMP or dAMP) and the templating 8-oxoG. The effect of this stacking on the ability of hPolβ to extend from 8-oxoG during long-patch base excision repair was unknown. Here we report pre-steady-state kinetics and time-dependent crystal structures to demonstrate that extension from both 8-oxoG:dC and 8-oxoG:dA base pairs is 18- to 580-fold less efficient compared to 8-oxoG bypass and that extension from 8-oxoG:dC over 8-oxoG:dA is favored by 15-fold. The overall decrease in efficiency of extension relative to 8-oxoG bypass is due to an alternative nucleotide binding conformation in the precatalytic ternary structures (hPolβ·DNA·dNTP) for both extension contexts, wherein the incoming nucleotide is bound in either the canonical Watson-Crick base pair or a nonplanar base pair. In addition, the decreased stability of the ternary complex of 8-oxoG:dA extension results in further loss of efficiency when compared to 8-oxoG:dC extension. Therefore, we hypothesize that the inefficient extension from 8-oxoG:dA serves as a newly discovered fidelity checkpoint during base excision repair.
- Department of Chemistry and Biochemistry, The Ohio State Biochemistry Program, The Ohio State University , Columbus, Ohio 43210, United States.
Organizational Affiliation: