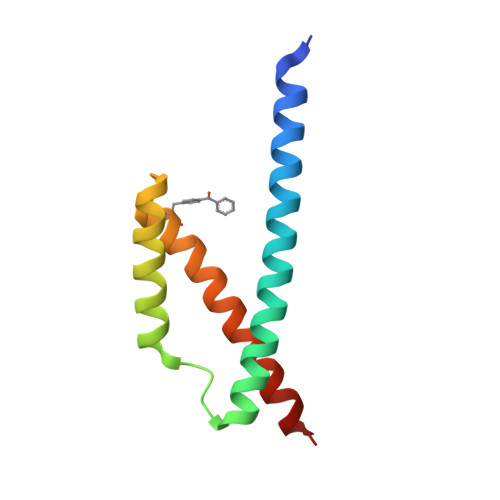
Generation of an Orthogonal Protein-Protein Interface with a Noncanonical Amino Acid.
Koh, M., Nasertorabi, F., Han, G.W., Stevens, R.C., Schultz, P.G.(2017) J Am Chem Soc 139: 5728-5731
- PubMed: 28413876
- DOI: https://doi.org/10.1021/jacs.7b02273
- Primary Citation of Related Structures:
5VHT - PubMed Abstract:
We have engineered the protein interface of the Escherichia coli chorismate mutase (EcCM) homodimer to be dependent on incorporation of a noncanonical amino acid (ncAA) at residue 72. The large hydrophobic amino acid p-benzoyl phenylalanine (pBzF) was substituted for Tyr72, which led to a catalytically inactive protein. A library of five residues (Leu25', Arg29', Leu76, Ile80' and Asp83') surrounding pBzF72 was generated and subjected to a growth based selection in a chorismate mutase deficient strain. An EcCM variant (Phe25', pBzF72, Thr76, Gly80' and Tyr83') forms a stable homodimer, has catalytic activity similar to the wild type enzyme, and unfolds with a T m of 53 °C. The X-ray crystal structure reveals a pi-pi stacking and hydrogen bonding interactions that stabilize the new protein interface. The strategy described here should be useful for generating organisms that are dependent on the presence of a ncAA for growth.
- Department of Chemistry and Skaggs Institute for Chemical Biology, The Scripps Research Institute , 10550 N Torrey Pines Road, La Jolla, California 92037, United States.
Organizational Affiliation: