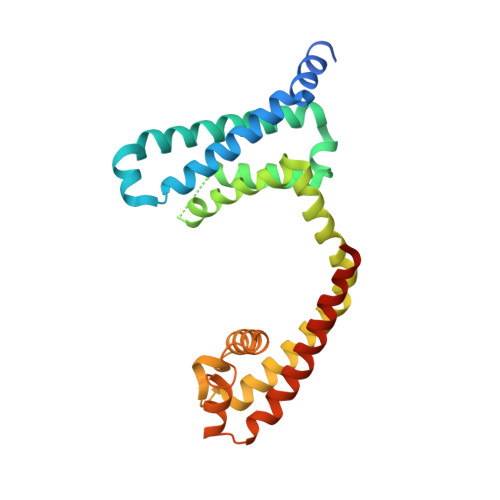
Structures of closed and open states of a voltage-gated sodium channel.
Lenaeus, M.J., Gamal El-Din, T.M., Ing, C., Ramanadane, K., Pomes, R., Zheng, N., Catterall, W.A.(2017) Proc Natl Acad Sci U S A 114: E3051-E3060
- PubMed: 28348242
- DOI: https://doi.org/10.1073/pnas.1700761114
- Primary Citation of Related Structures:
5VB2, 5VB8 - PubMed Abstract:
Bacterial voltage-gated sodium channels (BacNavs) serve as models of their vertebrate counterparts. BacNavs contain conserved voltage-sensing and pore-forming domains, but they are homotetramers of four identical subunits, rather than pseudotetramers of four homologous domains. Here, we present structures of two Na V Ab mutants that capture tightly closed and open states at a resolution of 2.8-3.2 Å. Introduction of two humanizing mutations in the S6 segment (Na V Ab/FY: T206F and V213Y) generates a persistently closed form of the activation gate in which the intracellular ends of the four S6 segments are drawn tightly together to block ion permeation completely. This construct also revealed the complete structure of the four-helix bundle that forms the C-terminal domain. In contrast, truncation of the C-terminal 40 residues in Na v Ab/1-226 captures the activation gate in an open conformation, revealing the open state of a BacNav with intact voltage sensors. Comparing these structures illustrates the full range of motion of the activation gate, from closed with its orifice fully occluded to open with an orifice of ∼10 Å. Molecular dynamics and free-energy simulations confirm designation of Na V Ab/1-226 as an open state that allows permeation of hydrated Na + , and these results also support a hydrophobic gating mechanism for control of ion permeation. These two structures allow completion of a closed-open-inactivated conformational cycle in a single voltage-gated sodium channel and give insight into the structural basis for state-dependent binding of sodium channel-blocking drugs.
- Department of Pharmacology, University of Washington, Seattle, WA 98195.
Organizational Affiliation: