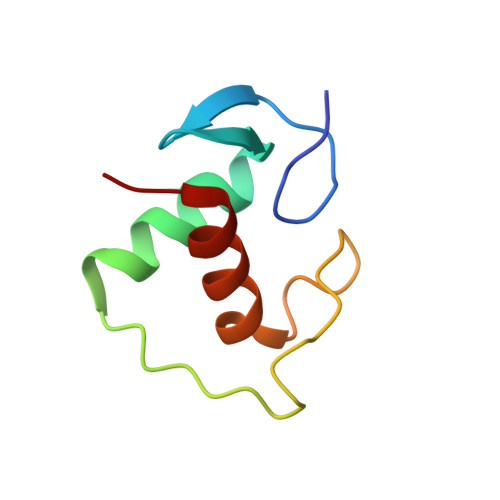
Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes.
Weikum, E.R., de Vera, I.M.S., Nwachukwu, J.C., Hudson, W.H., Nettles, K.W., Kojetin, D.J., Ortlund, E.A.(2017) Nucleic Acids Res 45: 8596-8608
- PubMed: 28591827
- DOI: https://doi.org/10.1093/nar/gkx509
- Primary Citation of Related Structures:
5VA0, 5VA7 - PubMed Abstract:
The glucocorticoid receptor (GR) is a ligand-regulated transcription factor that controls the expression of extensive gene networks, driving both up- and down-regulation. GR utilizes multiple DNA-binding-dependent and -independent mechanisms to achieve context-specific transcriptional outcomes. The DNA-binding-independent mechanism involves tethering of GR to the pro-inflammatory transcription factor activator protein-1 (AP-1) through protein-protein interactions. This mechanism has served as the predominant model of GR-mediated transrepression of inflammatory genes. However, ChIP-seq data have consistently shown GR to occupy AP-1 response elements (TREs), even in the absence of AP-1. Therefore, the current model is insufficient to explain GR action at these sites. Here, we show that GR regulates a subset of inflammatory genes in a DNA-binding-dependent manner. Using structural biology and biochemical approaches, we show that GR binds directly to TREs via sequence-specific contacts to a GR-binding sequence (GBS) half-site found embedded within the TRE motif. Furthermore, we show that GR-mediated transrepression observed at TRE sites to be DNA-binding-dependent. This represents a paradigm shift in the field, showing that GR uses multiple mechanisms to suppress inflammatory gene expression. This work further expands our understanding of this complex multifaceted transcription factor.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Organizational Affiliation: