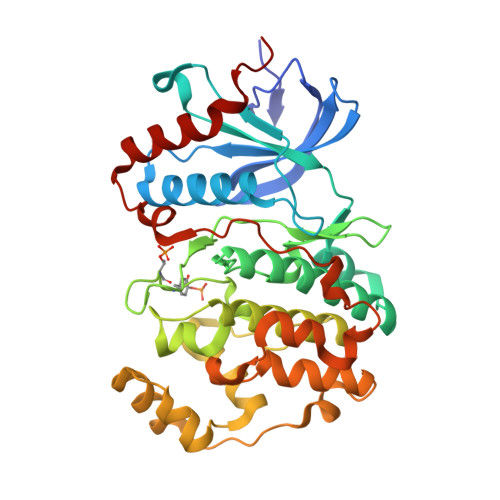
Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
Lechtenberg, B.C., Mace, P.D., Sessions, E.H., Williamson, R., Stalder, R., Wallez, Y., Roth, G.P., Riedl, S.J., Pasquale, E.B.(2017) ACS Med Chem Lett 8: 726-731
- PubMed: 28740606
- DOI: https://doi.org/10.1021/acsmedchemlett.7b00127
- Primary Citation of Related Structures:
5V60, 5V61, 5V62 - PubMed Abstract:
ERK is the effector kinase of the RAS-RAF-MEK-ERK signaling cascade, which promotes cell transformation and malignancy in many cancers and is thus a major drug target in oncology. Kinase inhibitors targeting RAF or MEK are already used for the treatment of certain cancers, such as melanoma. Although the initial response to these drugs can be dramatic, development of drug resistance is a major challenge, even with combination therapies targeting both RAF and MEK. Importantly, most resistance mechanisms still rely on activation of the downstream effector kinase ERK, making it a promising target for drug development efforts. Here, we report the design and structural/functional characterization of a set of bivalent ERK inhibitors that combine a small molecule inhibitor that binds to the ATP-binding pocket with a peptide that selectively binds to an ERK protein interaction surface, the D-site recruitment site (DRS). Our studies show that the lead bivalent inhibitor, SBP3, has markedly improved potency compared to the small molecule inhibitor alone. Unexpectedly, we found that SBP3 also binds to several ERK-related kinases that contain a DRS, highlighting the importance of experimentally verifying the predicted specificity of bivalent inhibitors. However, SBP3 does not target any other kinases belonging to the same CMGC branch of the kinome. Additionally, our modular click chemistry inhibitor design facilitates the generation of different combinations of small molecule inhibitors with ERK-targeting peptides.
- Cancer Center, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, California 92037, United States.
Organizational Affiliation: