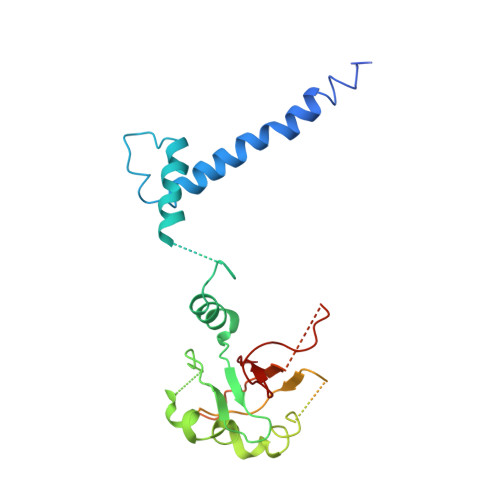
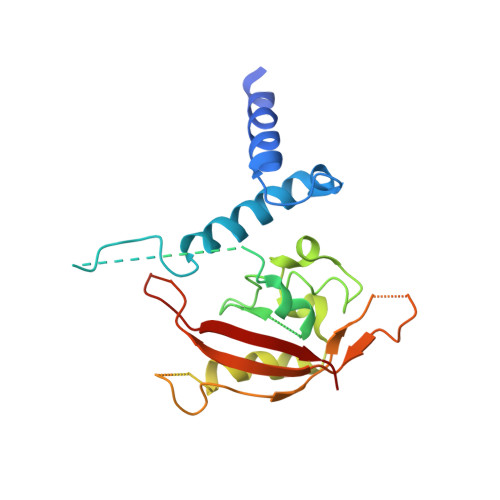
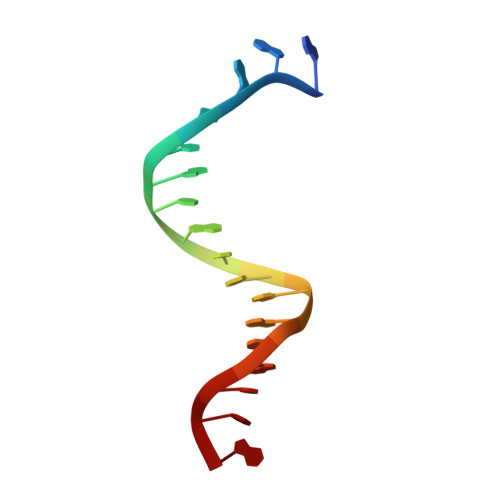
Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex.
Seok, S.H., Lee, W., Jiang, L., Molugu, K., Zheng, A., Li, Y., Park, S., Bradfield, C.A., Xing, Y.(2017) Proc Natl Acad Sci U S A 114: 5431-5436
- PubMed: 28396409
- DOI: https://doi.org/10.1073/pnas.1617035114
- Primary Citation of Related Structures:
5V0L - PubMed Abstract:
The aryl hydrocarbon receptor (AHR) belongs to the PAS (PER-ARNT-SIM) family transcription factors and mediates broad responses to numerous environmental pollutants and cellular metabolites, modulating diverse biological processes from adaptive metabolism, acute toxicity, to normal physiology of vascular and immune systems. The AHR forms a transcriptionally active heterodimer with ARNT (AHR nuclear translocator), which recognizes the dioxin response element (DRE) in the promoter of downstream genes. We determined the crystal structure of the mammalian AHR-ARNT heterodimer in complex with the DRE, in which ARNT curls around AHR into a highly intertwined asymmetric architecture, with extensive heterodimerization interfaces and AHR interdomain interactions. Specific recognition of the DRE is determined locally by the DNA-binding residues, which discriminates it from the closely related hypoxia response element (HRE), and is globally affected by the dimerization interfaces and interdomain interactions. Changes at the interdomain interactions caused either AHR constitutive nuclear localization or failure to translocate to nucleus, underlying an allosteric structural pathway for mediating ligand-induced exposure of nuclear localization signal. These observations, together with the global higher flexibility of the AHR PAS-A and its loosely packed structural elements, suggest a dynamic structural hierarchy for complex scenarios of AHR activation induced by its diverse ligands.
- McArdle Laboratory for Cancer Research, Department of Oncology, School of Medicine and Public Health, University of Wisconsin, Madison, WI 53705.
Organizational Affiliation: