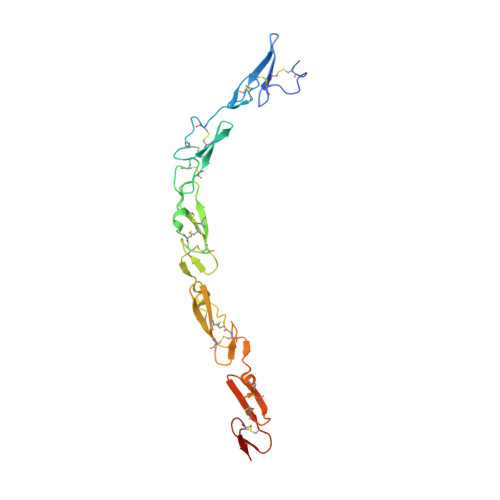
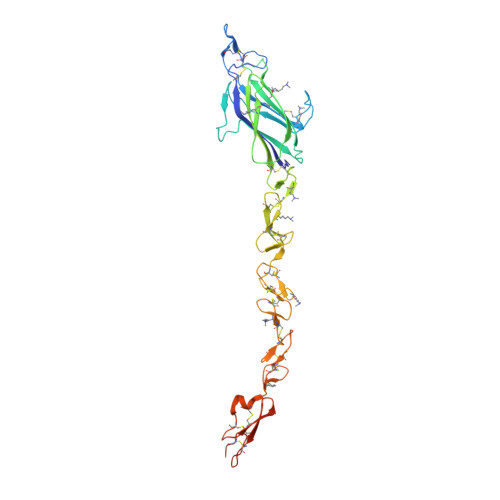
Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity.
Luca, V.C., Kim, B.C., Ge, C., Kakuda, S., Wu, D., Roein-Peikar, M., Haltiwanger, R.S., Zhu, C., Ha, T., Garcia, K.C.(2017) Science 355: 1320-1324
- PubMed: 28254785
- DOI: https://doi.org/10.1126/science.aaf9739
- Primary Citation of Related Structures:
5UK5 - PubMed Abstract:
Notch receptor activation initiates cell fate decisions and is distinctive in its reliance on mechanical force and protein glycosylation. The 2.5-angstrom-resolution crystal structure of the extracellular interacting region of Notch1 complexed with an engineered, high-affinity variant of Jagged1 (Jag1) reveals a binding interface that extends ~120 angstroms along five consecutive domains of each protein. O -Linked fucose modifications on Notch1 epidermal growth factor-like (EGF) domains 8 and 12 engage the EGF3 and C2 domains of Jag1, respectively, and different Notch1 domains are favored in binding to Jag1 than those that bind to the Delta-like 4 ligand. Jag1 undergoes conformational changes upon Notch binding, exhibiting catch bond behavior that prolongs interactions in the range of forces required for Notch activation. This mechanism enables cellular forces to regulate binding, discriminate among Notch ligands, and potentiate Notch signaling.
- Departments of Molecular and Cellular Physiology and Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: