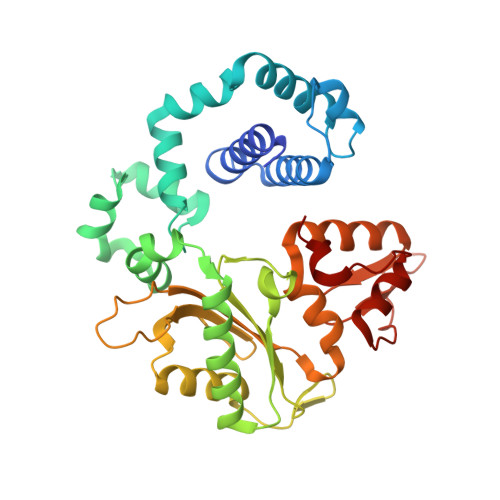
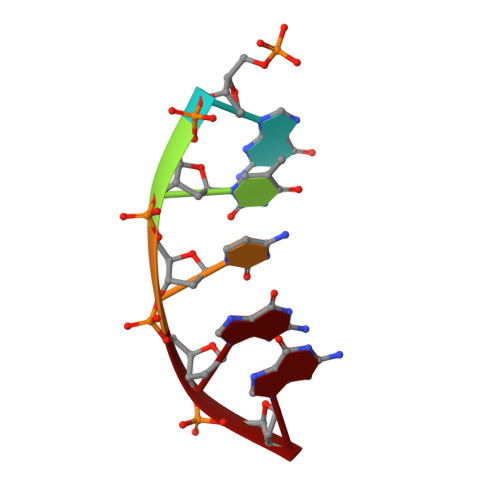
Revealing the role of the product metal in DNA polymerase beta catalysis.
Perera, L., Freudenthal, B.D., Beard, W.A., Pedersen, L.G., Wilson, S.H.(2017) Nucleic Acids Res 45: 2736-2745
- PubMed: 28108654
- DOI: https://doi.org/10.1093/nar/gkw1363
- Primary Citation of Related Structures:
5U9H - PubMed Abstract:
DNA polymerases catalyze a metal-dependent nucleotidyl transferase reaction during extension of a DNA strand using the complementary strand as a template. The reaction has long been considered to require two magnesium ions. Recently, a third active site magnesium ion was identified in some DNA polymerase product crystallographic structures, but its role is not known. Using quantum mechanical/ molecular mechanical calculations of polymerase β, we find that a third magnesium ion positioned near the newly identified product metal site does not alter the activation barrier for the chemical reaction indicating that it does not have a role in the forward reaction. This is consistent with time-lapse crystallographic structures following insertion of Sp-dCTPαS. Although sulfur substitution deters product metal binding, this has only a minimal effect on the rate of the forward reaction. Surprisingly, monovalent sodium or ammonium ions, positioned in the product metal site, lowered the activation barrier. These calculations highlight the impact that an active site water network can have on the energetics of the forward reaction and how metals or enzyme side chains may interact with the network to modulate the reaction barrier. These results also are discussed in the context of earlier findings indicating that magnesium at the product metal position blocks the reverse pyrophosphorolysis reaction.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, P.O. Box 12233, Research Triangle Park, NC 27709-2233, USA.
Organizational Affiliation: