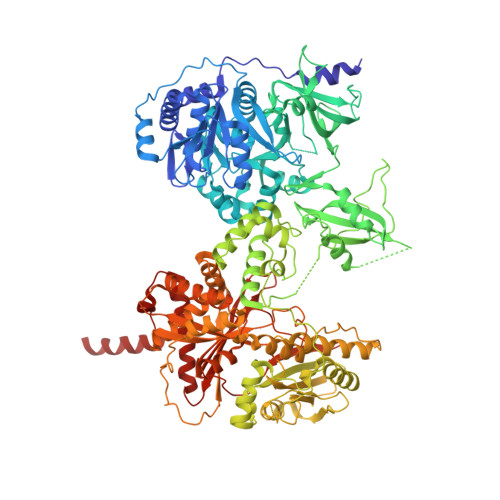
X-Ray Crystallography and Electron Microscopy of Cross- and Multi-Module Nonribosomal Peptide Synthetase Proteins Reveal a Flexible Architecture.
Tarry, M.J., Haque, A.S., Bui, K.H., Schmeing, T.M.(2017) Structure 25: 783-793.e4
- PubMed: 28434915
- DOI: https://doi.org/10.1016/j.str.2017.03.014
- Primary Citation of Related Structures:
5U89 - PubMed Abstract:
Nonribosomal peptide synthetases (NRPS) are macromolecular machines that produce peptides with diverse activities. Structural information exists for domains, didomains, and even modules, but little is known about higher-order organization. We performed a multi-technique study on constructs from the dimodular NRPS DhbF. We determined a crystal structure of a cross-module construct including the adenylation (A) and peptidyl carrier protein (PCP) domains from module 1 and the condensation domain from module 2, complexed with an adenosine-vinylsulfonamide inhibitor and an MbtH-like protein (MLP). The action of the inhibitor and the role of the MLP were investigated using adenylation reactions and isothermal titration calorimetry. In the structure, the PCP and A domains adopt a novel conformation, and noncovalent, cross-module interactions are limited. We calculated envelopes of dimodular DhbF using negative-stain electron microscopy. The data show large conformational variability between modules. Together, our results suggest that NRPSs lack a uniform, rigid supermodular architecture.
- Department of Biochemistry, McGill University, Montréal, QC H3G 0B1, Canada.
Organizational Affiliation: