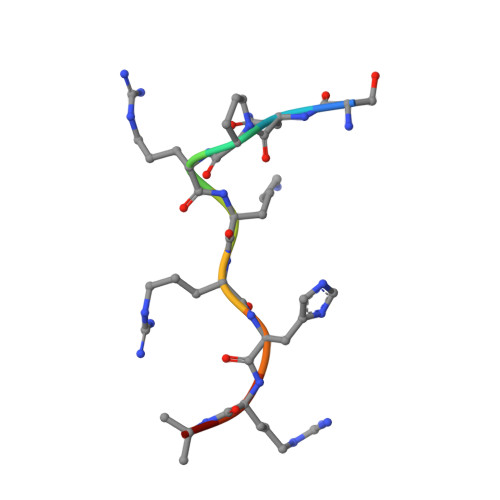
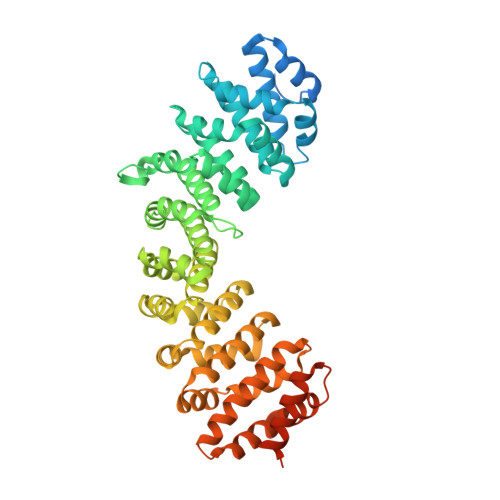
DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
de Barros, A.C., Takeda, A.A.S., Dreyer, T.R., Velazquez-Campoy, A., Kobe, B., Fontes, M.R.M.(2018) Biochimie 146: 87-96
- PubMed: 29175432
- DOI: https://doi.org/10.1016/j.biochi.2017.11.013
- Primary Citation of Related Structures:
5U5P, 5U5R - PubMed Abstract:
MLH1 and PMS2 proteins form the MutLα heterodimer, which plays a major role in DNA mismatch repair (MMR) in humans. Mutations in MMR-related proteins are associated with cancer, especially with colon cancer. The N-terminal region of MutLα comprises the N-termini of PMS2 and MLH1 and, similarly, the C-terminal region of MutLα is composed by the C-termini of PMS2 and MLH1, and the two are connected by linker region. The nuclear localization sequences (NLSs) necessary for the nuclear transport of the two proteins are found in this linker region. However, the exact NLS sequences have been controversial, with different sequences reported, particularly for MLH1. The individual components are not imported efficiently, presumably due to their C-termini masking their NLSs. In order to gain insights into the nuclear transport of these proteins, we solved the crystal structures of importin-α bound to peptides corresponding to the supposed NLSs of MLH1 and PMS2 and performed isothermal titration calorimetry to study their binding affinities. Both putative MLH1 and PMS2 NLSs can bind to importin-α as monopartite NLSs, which is in agreement with some previous studies. However, MLH1-NLS has the highest affinity measured by a natural NLS peptide, suggesting a major role of MLH1 protein in nuclear import compared to PMS2. Finally, the role of MLH1 and PMS2 in the nuclear transport of the MutLα heterodimer is discussed.
- Departamento de Física e Biofísica, Instituto de Biociências, Universidade Estadual Paulista, Botucatu, SP, Brazil.
Organizational Affiliation: