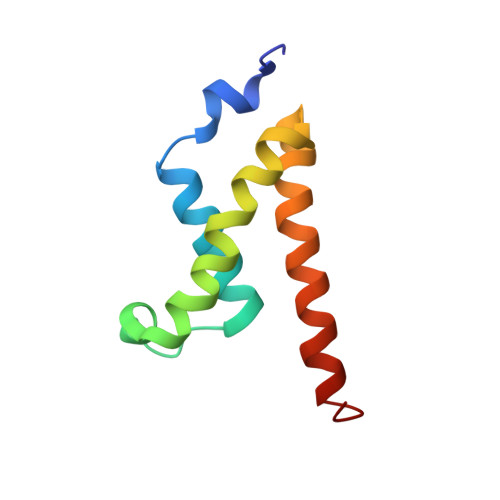
Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Lecoq, L., Raiola, L., Chabot, P.R., Cyr, N., Arseneault, G., Legault, P., Omichinski, J.G.(2017) Nucleic Acids Res 45: 5564-5576
- PubMed: 28334776
- DOI: https://doi.org/10.1093/nar/gkx146
- Primary Citation of Related Structures:
5U4K, 5URN - PubMed Abstract:
p65 is a member of the NF-κB family of transcriptional regulatory proteins that functions as the activating component of the p65-p50 heterodimer. Through its acidic transactivation domain (TAD), p65 has the capacity to form interactions with several different transcriptional regulatory proteins, including TFIIB, TFIIH, CREB-binding protein (CBP)/p300 and TAFII31. Like other acidic TADs, the p65 TAD contains two subdomains (p65TA1 and p65TA2) that interact with different regulatory factors depending on the target gene. Despite its role in controlling numerous NF-κB target genes, there are no high-resolution structures of p65TA1 bound to a target transcriptional regulatory factor. In this work, we characterize the interaction of p65TA1 with two factors, the Tfb1/p62 subunit of TFIIH and the KIX domain of CBP. In these complexes, p65TA1 transitions into a helical conformation that includes its characteristic ΦXXΦΦ motif (Φ = hydrophobic amino acid). Structural and functional studies demonstrate that the two binding interfaces are primarily stabilized by three hydrophobic amino acids within the ΦXXΦΦ motif and these residues are also crucial to its ability to activate transcription. Taken together, the results provide an atomic level description of how p65TA1 is able to bind different transcriptional regulatory factors needed to activate NF-κB target genes.
- Department of Biochemistry and Molecular Medicine, Université de Montréal, Montréal, QC H3C 3J7, Canada.
Organizational Affiliation: